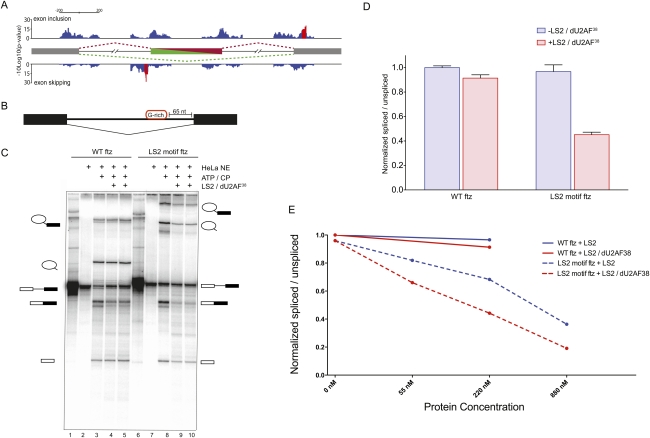
Figure 5.
LS2 inhibits splicing of the ftz intron in vitro. (A) Motif location and clustering in LS2-affected cassette exons. LS2-affected cassette exons were searched for LS2 recognition motifs in 50-nt overlapping windows. Motif-containing windows were called as significant (red bars) if they contained a significant motif enrichment (P-value < 0.05) and were part of a stretch of at least five consecutive significant windows. LS2 recognition motifs associated with cassette exon inclusion are displayed on top, while motifs associated with cassette exon skipping are on the bottom. (B) Diagram of the modified ftz intron in vitro splicing substrate generated with and without a G-rich LS2 recognition motif inserted 65 nt upstream of the 3′ splice site. (C) In vitro splicing reaction of the wild-type (lanes 1–5) and LS2 SELEX motif-containing (lanes 6–10) ftz substrates using HeLa cell nuclear extract in the presence or absence of purified recombinant GST-tagged LS2/dU2AF38 heterodimer. The identity of each RNA species is shown schematically at the right and left of the panel. (CP) Creatine phosphate. Reactions were carried out without HeLa nuclear extract (lanes 1,6); with nuclear extract but without ATP and CP (lanes 2,7); with nuclear extract, ATP, and CP (lanes 3,8); and with nuclear extract, ATP, CP, and 500 ng of recombinant LS2/dU2AF38 heterodimer protein (lanes 4,5,9,10). (D) PhosphorImager quantification of the results in B. The Y-axis represents the ratio of all splicing intermediate species to the unspliced pre-mRNA, with intensities for each species normalized to their length and all ratios normalized such that the value for wild-type (WT) ftz without added LS2/dU2AF38 heterodimer is 1.0. Error bars represent standard deviations of four to six experiments. (E) In vitro splicing efficiency of wild-type (WT) ftz and LS2 motif ftz in the presence of varying amounts of purified recombinant LS2 and LS2/dU2AF38 heterodimer. Quantification was performed as in D.