Abstract
Recently, many studies have reported that polyamines play a role in bacterial cell-to-cell signaling processes. The present study describes a novel putrescine importer required for induction of type 1 pili-driven surface motility. The surface motility of the Escherichia coli ΔspeAB ΔspeC ΔpotABCD strain, which cannot produce putrescine and cannot import spermidine from the medium, was induced by extracellular putrescine. Introduction of the gene deletions for known polyamine importers (ΔpotE, ΔpotFGHI, and ΔpuuP) or a putative polyamine importer (ΔydcSTUV) into the ΔspeAB ΔspeC ΔpotABCD strain did not affect putrescine-induced surface motility. The deletion of yeeF, an annotated putative putrescine importer, in the ΔspeAB ΔspeC ΔpotABCD ΔydcSTUV strain abolished surface motility in putrescine-supplemented medium. Complementation of yeeF by a plasmid vector restored surface motility. The surface motility observed in the present study was abolished by the deletion of fimA, suggesting that the surface motility is type 1 pili-driven. A transport assay using the yeeF+ or ΔyeeF strains revealed that YeeF is a novel putrescine importer. The Km of YeeF (155 μm) is 40 to 300 times higher than that of other importers reported previously. On the other hand, the Vmax of YeeF (9.3 nmol/min/mg) is comparable to that of PotABCD, PotFGHI, and PuuP. The low affinity of YeeF for putrescine may allow E. coli to sense the cell density depending on the concentration of extracellular putrescine.
Keywords: Bacteria, Bacterial Signal Transduction, Cell Motility, Polyamines, Transport Amino Acids, Escherichia coli, Cell-to-Cell Signaling, Pili, Putrescine, Transporter
Introduction
Polyamines are aliphatic amines with two or more amino groups in their chemical structure. Basic polyamines are putrescine, spermidine, and spermine. In addition, a variety of unusual polyamines has been reported in several plants (1) and bacteria (2). Polyamines are widely distributed from prokaryotic (3) to eukaryotic cells (4) and are found at especially high concentrations in proliferating cells such as cancer cells (5) and bacteria in the exponential growth phase (6). Polyamines are important growth factors because they stabilize nucleic acid structure (7) and promote protein synthesis (8).
Putrescine is synthesized in Escherichia coli cells from ornithine by ornithine decarboxylase (SpeC) or from arginine by the sequential actions of arginine decarboxylase (SpeA) and agmatinase (SpeB) (9). Putrescine is converted to spermidine, another polyamine, by the addition of an aminopropyl group derived from decarboxylated S-adenosylmethionine by spermidine synthase (SpeE) (10).
Recently, many studies have shown that polyamines play a role in cell-to-cell signaling processes in bacteria. In Yersinia pestis, the etiological agent of bubonic and pneumonic plague, the components of the putrescine synthetic pathway, SpeA and SpeC are essential for the formation of normal biofilms (11). Furthermore, ΔspeA ΔspeC mutants can only form biofilms when the medium is supplemented with putrescine (11). In Proteus mirabilis, a common human urinary tract pathogen, putrescine acts as a cell-to-cell signaling molecule that induces differentiation into swarm cells (12). When extracellular polyamines are used as signaling molecules, bacteria require a polyamine sensor or polyamine importer because polyamines are hydrophilic molecules that cannot passively cross the plasma membrane. In Vibrio cholerae, a human intestinal pathogen, the unusual polyamine norspermidine activates biofilm formation in a norspermidine sensor NspS-dependent manner (13). In E. coli, spermidine induces surface motility in a spermidine importer PotABCD-dependent manner (14).
In E. coli, four polyamine importers have been identified. PotFGHI (15) has been identified as an ATP-dependent putrescine transporter of the ABC transporter family. PotABCD is a spermidine transporter of the ABC transporter family that takes up putrescine with lower affinity (16, 17). PotE is responsible for both the excretion and uptake of putrescine (18). PuuP is the most recently discovered putrescine transporter and depends on proton motive force (19, 20). In addition, two putative putrescine importers, YdcSTUV and YeeF, have been annotated by computer analysis.
In this paper, we report a novel putrescine importer, YeeF that was identified using mutants with multiple deletions of genes involved in polyamine synthesis and transport. YeeF was indispensable for surface motility induced by extracellular putrescine as a signaling molecule.
EXPERIMENTAL PROCEDURES
Strain and Plasmid Construction
The 2869-bp fragments, including the entire region of yeeFE, the 14-bp region downstream of yeeFE and the 260-bp region upstream of yeeFE, were amplified by PCR using the genome of SH639 as template (Table 1). KOD-plus-DNA polymerase (Toyobo, Osaka, Japan) and the primers “yeeF up EcoRI” and “yeeE down NcoI” were used to perform PCR according to the manufacturer's instructions. The primers were designed to add EcoRI and NcoI restriction sites to the 5′ and 3′ ends of the amplified region, respectively. The amplified fragment was ligated to a 3.9-kb EcoRI and NcoI fragment of pACYC184, and this plasmid was designated pACYC184-yeeF+E+. The cloned region containing yeeFE was sequenced to confirm that no mutations had occurred. pACYC184-yeeF+E+ was digested with ScaI, and the obtained 6-kb fragment was self-ligated. This plasmid was designated pACYC184-yeeF+. pACYC184-yeeF+E+ was digested with EcoRI and ScaI and blunt-ended with a DNA blunting kit (Takara, Otsu, Japan). The obtained 2.2-kb fragment containing the yeeF gene was ligated to HpaI-digested pBelobac11. This plasmid was designated pBelobac11-yeeF+. The 1049-bp fragment, including the entire region of fimA and the 500-bp region upstream of fimA, was amplified by PCR using the genome of SH639 as a template. KOD-plus-DNA polymerase (Toyobo) and the primers CCC-EcoRV-fimA_upstream and CCC-SphI-fimA_downstream were used to perform the PCR, according to the manufacturer's instructions. The primers were designed to add EcoRV and SphI restriction sites to the 5′ and 3′ ends of the amplified region, respectively. The amplified fragment was ligated to a 3.9-kb EcoRV and SphI fragment of pACYC184, and this plasmid was designated pACYC184-fimA+. The region containing fimA and fimA-promoter was sequenced to confirm that no mutations had occurred. Gene disruption of speAB, speC, potABCD, potE, potFGHI, ydcSTUV, yeeF, fliC, and fimA was performed by the method described previously (21). Gene disruption of puuP was performed as described previously (20). P1 transduction (22) was used to construct the multiple mutants or transfer the chromosomal deletion of a gene into another strain.
TABLE 1.
Strains used in this study
| Strain, plasmid, and oligonucleotide | Characteristic or sequence | Source or reference |
|---|---|---|
| Strain | ||
| JW1908 | ME9062 except ΔfliC::FRT-kan+-FRT | Ref. 36 |
| JW4277 | ME9062 except ΔfimA::FRT-kan+-FRT | Ref. 36 |
| ME9062 | rrnB ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | Ref. 36 |
| MG1655 | F− prototrophic | C. A. Gross |
| SH639 | F− Δggt-2 | Refs. 37 and 38 |
| SK430 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT-cat+-FRT | This study |
| SK454 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT-cat+-FRT ΔydcSTUV::FRT-kan+-FRT | This study |
| SK468 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT-cat+-FRT ΔpotE::FRT-kan+-FRT | This study |
| SK469 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT-cat+-FRT ΔpuuP:: tet+ | This study |
| SK479 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT | Ref. 14 |
| SK489 | pBelobac11/YT16 | Ref. 14 |
| SK496 | MG1655 except ΔyeeF::FRT-cat+-FRT | This study |
| SK505 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT ΔyeeF::FRT-cat+-FRT | This study |
| SK531 | pBelobac11/SK479 | Ref. 14 |
| SK560 | pBelobac11-potA+B+C+D+/SK479 | Ref. 14 |
| SK580 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT ΔydcSTUV::FRT | This study |
| SK582 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT ΔydcSTUV::FRT ΔyeeF::FRT | This study |
| SK591 | pBelobac11/SK580 | This study |
| SK594 | pBelobac11-yeeF+/SK582 | This study |
| SK595 | pBelobac11/SK582 | This study |
| SK598 | MG1655 except ΔfliC::FRT-kan+-FRT | This study |
| SK624 | SH639 except ΔpotABCD::FRT ΔpotE::FRT ΔpotFGHI::FRT ΔpuuP::FRT-kan+-FRT ΔydcSTUV::FRT ΔyeeF::FRT | This study |
| SK629 | pACYC184-yeeF+/SK624 | This study |
| SK644 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT ΔydcSTUV::FRT ΔfimA::FRT-kan+-FRT | This study |
| SK650 | MG1655 except ΔfimA:: FRT-kan+-FRT | This study |
| SK651 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT ΔydcSTUV::FRT ΔfliC::FRT-kan+-FRT | This study |
| SK653 | pACYC184/SK580 | This study |
| SK654 | pACYC184/SK644 | This study |
| SK655 | pACYC184-fimA+/SK644 | This study |
| YT16 | SH639 except ΔspeAB::FRT ΔspeC::FRT | Ref. 14 |
| YT21 | SH639 except ΔspeAB::FRT ΔspeC::FRT ΔpotABCD::FRT-cat+-FRT | This study |
| ΔpotFGHI::FRT-kan+-FRT | ||
| Plasmid | ||
| pACYC184 | p15A replicon cat+tet+ | New England Biolabs |
| pACYC184-fimA+ | p15A replicon cat+fimA+ | This study |
| pACYC184-yeeF+ | p15A replicon tet+yeeF+ | This study |
| pACYC184-yeeF+E+ | p15A replicon tet+yeeF+E+ | This study |
| pBelobac11 | Mini-F replicon cat+ | New England Biolabs |
| pBelobac11-yeeF+ | Mini-F replicon cat+yeeF+ | This study |
| pBelobac11-potA+B+C+D+ | Mini-F replicon cat+potA+B+C+D+ | Ref. 14 |
| Oligonucleotide | ||
| CCC-EcoRV-fimA_upstream | 5′-CCCGATATCAAAAAGAGAAGAGGTTTGAT-3′ | |
| CCC-SphI-fimA_downstream | 5′-CCCGCATGCTTATTGATACTGAACCTTGA-3′ | |
| yeeF up EcoRI | 5′-CCCGAATTCATTATTCTGACAAGCCTCTC-3′ | |
| yeeE down NcoI | 5′-CCCCCATGGTCTTTACACCTTACTTAATT-3′ | |
Media and Culture Conditions
LB glucose semisolid (LBGS)2 plates were prepared with LB (BD, Franklin Lakes, NJ), 0.5% glucose, and 0.6% agar (Eiken, Tokyo, Japan). LB glucose (LBG) is the liquid medium containing LB and 0.5% glucose. Chloramphenicol (15 μg/ml) was supplemented to the medium to maintain the plasmids. In experiments in which surface motility was evaluated, cultures were diluted in LB to an A600 of 0.4, and 3 μl of the diluted culture were spotted onto the center of LBGS, supplemented or not with 400 μm putrescine, and incubated at 37 °C. Forty h after inoculation, the plates were photographed.
DNA Microarray Analysis
DNA microarray was performed using the E. coli_K12 Expr 4 × 72K Array Service (Roche Applied Science) which contains 72,000 oligonucleotide probes corresponding to ∼4,800 E. coli genes. The probe length is 45- to 60-mer, and the number of probes per gene is 6 to 22.
Functional Gene Pathway Analysis (PAGE)
The microarray data were further analyzed by a PAGE analysis (23) using gene ontology terms described in a profiling of E. coli chromosome (PEC) database (24). Gene ontology terms are ranked by p value.
Transport Assay
The transport assay was performed using [14C]putrescine dihydrochloride as described previously (25), with the difference that M9-glucose medium was used instead of M63 medium. The disintegrations per minute of 14C in the cell were divided by the protein amount of the cell used in the transport assay. The cell protein content was extracted as described previously (26), and the amount of protein was assayed by Lowry's method (27).
Analysis of Polyamine Concentration
The concentration of polyamines in the samples was measured as described previously (26).
RESULTS
Putrescine-induced Surface Motility Is Independent of potABCD
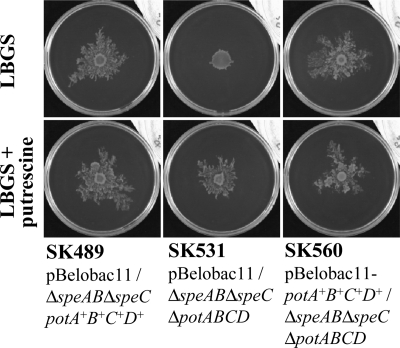
In a previous study (14), we reported that the surface motility of strains with a ΔspeAB ΔspeC genetic background, depends on the spermidine importer PotABCD and spermidine, which had been originally included in LBGS plates (Fig. 1). It has been reported that PotABCD can take up not only spermidine but also putrescine (17). However, when 400 μm putrescine were added to the medium, surface motility was PotABCD-independent (Fig. 1). This result suggested that other putrescine importers could be responsible for induction of surface motility by extracellular putrescine.
FIGURE 1.
Surface motility is induced by extracellular putrescine. Bacterial cultures were diluted in LB medium to an A600 of 0.4, and 3 μl of the diluted cultures were dropped onto the center of LBGS plates, supplemented with or without 400 μm putrescine, and incubated at 37 °C. The plates were photographed 40 h after inoculation.
Search for a Putrescine Importer Responsible for Putrescine-induced Surface Motility
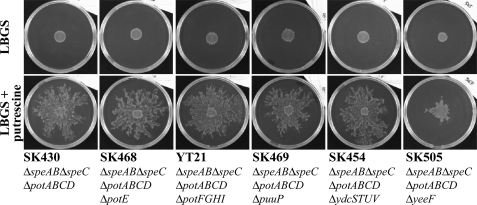
To determine the importer responsible for putrescine-induced surface motility, gene deletions for putrescine importers or putative putrescine importers were introduced into a strain with a ΔspeAB ΔspeC ΔpotABCD genetic background (SK430). The genotypes of strains used in this study are shown in Table 1. The introduction of ΔpotE, ΔpotFGHI, ΔpuuP, or ΔydcSTUV into the ΔspeAB ΔspeC ΔpotABCD genetic background (strains SK468, YT21, SK496, and SK454) had no effect on surface motility on LBGS plates supplemented with 400 μm putrescine (Fig. 2). However, introduction of ΔyeeF into the ΔspeAB ΔspeC ΔpotABCD genetic background (strain SK505) resulted in severe inhibition of surface motility under the same conditions (Fig. 2). This result suggested that yeeF is the main factor responding to putrescine in the induction of surface motility of E. coli.
FIGURE 2.
The influence of gene disruptions of putrescine importers or putative putrescine importers on extracellular induced surface motility. Bacterial cultures were diluted in LB medium to an A600 of 0.4, and 3 μl of the diluted cultures were spotted onto the center of LBGS plates, supplemented with or without 400 μm putrescine, and incubated at 37 °C. The plates were photographed 40 h after inoculation.
Backup Role of ydcSTUV in Putrescine-induced Surface Motility
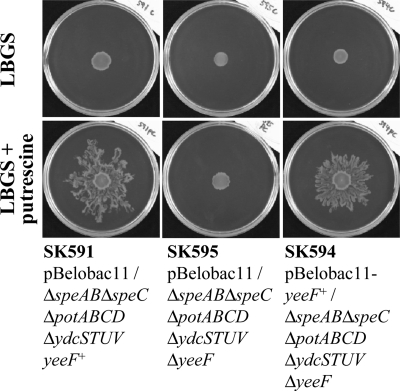
The surface motility of the ydcS+T+U+V+ΔspeAB ΔspeC ΔpotABCD ΔyeeF strain (SK505) on LBGS plates supplemented with 400 μm putrescine was reduced (Fig. 2), whereas that of the ΔydcSTUV ΔspeAB ΔspeC ΔpotABCD ΔyeeF strain (SK595) was completely abolished (Fig. 3). This result suggested that ydcSTUV plays a backup role in putrescine-induced surface motility. We are currently characterizing the role of ydcSTUV in more detail.
FIGURE 3.
The role of yeeF in surface motility induced by extracellular putrescine. Bacterial cultures were diluted in LB medium to an A600 of 0.4, and 3 μl of the diluted cultures were spotted onto the center of LBGS plates, supplemented with or without 400 μm putrescine, and incubated at 37 °C. The plates were photographed 40 h after inoculation. The medium was supplemented with chloramphenicol (15 μg/ml) to maintain the plasmids.
Role of yeeF in Putrescine-induced Surface Motility
To confirm whether yeeF plays a significant role in surface motility in response to putrescine, we performed a complementation experiment (Fig. 3). Both the yeeF+ strain (SK591) and the ΔyeeF strain complemented with a plasmid vector containing the yeeF+ gene (SK594) exhibited surface motility on LBGS plates supplemented with 400 μm putrescine, but not on putrescine-free LBGS plates. In contrast, the ΔyeeF strain (SK595) did not exhibit surface motility on LBGS plates, whether supplemented with 400 μm putrescine or not. This result clearly demonstrated that yeeF is an important factor in the induction of surface motility when extracellular putrescine is present in the environment.
Genome-wide Gene Expression Comparison between Strains That Exhibit Surface Motility and a Strain That Does Not
To search for genes regulated in cells that exhibited surface motility as compared with those that did not, we performed a DNA microarray using cDNA derived from RNA extracted from the yeeF+ (SK591), ΔyeeF (SK595), and pBelobac11-yeeF+/SK595 (SK594) strains cultured on LBGS plates supplemented with 400 μm putrescine. The expression of a significant number of pathways was altered when the gene expression profiles of SK591 or SK594 were compared with that of SK595 using PAGE analysis with that of SK595 (supplemental Tables S1–S4). The result of the comparison between SK591and SK595 (supplemental Tables S1 and S3) was very similar to that between SK594 and SK595 (supplemental Tables S2 and S4). This showed that the complementation of yeeF by the single-copy vector pBelobac11 in SK594 functioned properly. When the gene expression profiles of SK591 and SK594, which exhibit surface motility, were compared with that of SK595, which does not exhibit surface motility, many metabolic pathways of nutrient sources such as polyol, nitrate, and glycerol were significantly up-regulated (supplemental Tables S1 and S2). The expression of the flagellum organization, which is important for swarming motility, was also significantly up-regulated in SK591 and SK594 (Z score = 2.58 and 4.17, respectively) compared with SK595 (supplemental Tables S1 and S2). On the other hand, the synthetic pathways of arginine and amine as well as the stress response were down-regulated in SK591 and SK594 compared with SK595 (supplemental Table S3 and S4).
YeeF Is a Novel Putrescine Importer
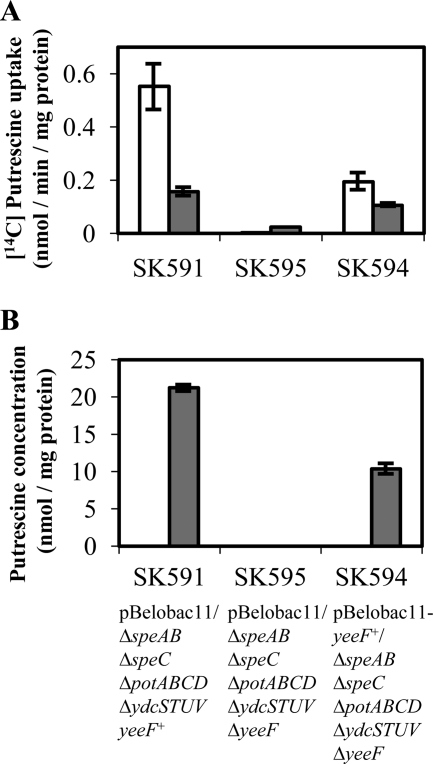
YeeF is a homologue (63% identity) of PuuP, a putrescine importer that we have previously discovered (20). YeeF has recently been annotated as a putrescine importer in several databases. In addition, the ΔyeeF mutant was listed in 294-single gene knock-out mutants that exhibited a repressed-swarming phenotype selected in a genome-wide screening (28). Yet, its function was not reported. To verify the putrescine uptake activity of YeeF, we performed a transport assay using [14C]putrescine and determined the concentration of putrescine in SK591, SK594, and SK595 cells grown in LBG with (Fig. 4, gray bars) or without (Fig. 4, white bars) 400 μm putrescine. The putrescine uptake activity of the yeeF+ strain (SK591) grown in putrescine-free LBG was 0.553 nmol/min/mg (Fig. 4A, white bar). The putrescine uptake activity of the ΔyeeF strain (SK595) grown in LBG without putrescine was not detected and that of the pBelobac11-yeeF+/SK595 strain (SK594) was 0.193 nmol/min/mg (Fig. 4A, white bars). One hundred μm carbonyl cyanide m-chlorophenylhydrazone completely abolished the activity of putrescine uptake in SK591 and SK594. Interestingly, the putrescine uptake activity of SK591 and SK594 grown in LBG with 400 μm putrescine was repressed to 28% (in SK591) and 55% (in SK594) compared with strains grown without putrescine (Fig. 4A, gray bars). However, intracellular putrescine was detected only in cells grown in LBG supplemented with 400 μm putrescine. The concentration of intracellular putrescine in SK591 and SK594 cells was 21.2 nmol/mg and 10.4 nmol/mg (Fig. 4B, gray bars). These values are converted to 7.3 mm (SK591) and 3.6 mm (SK594) based on a cell volume of 2.9 μl per mg protein (29). The intracellular putrescine was not detected in SK595. These results clearly showed that E. coli took up putrescine using YeeF, a novel putrescine importer.
FIGURE 4.
The uptake of extracellular putrescine depends on yeeF. Overnight precultures of SK591, SK595, and SK594 were inoculated in 100-ml LBG supplemented with (gray bars) or without (white bars) 400 μm putrescine in 500-ml Erlenmeyer flasks. The medium was supplemented with chloramphenicol (15 μg/ml) to maintain the plasmids. The initial optical density of the culture, measured at 600 nm, was adjusted to 0.001. The flasks were shaken at 150 rpm at 37 °C. Bacterial cells were harvested at A600 = 1.1–1.3 and washed once with cold M9 glucose. The assays were performed three times with independent bacterial cultures. Values are expressed as the mean ± S.D. A, [14C]putrescine uptake by strains. The cells were incubated with 50 μm [14C]putrescine, and the reaction was terminated after 0, 1, 7, 13, or 20 min. The linearity between the uptake of [14C]putrescine and the incubation time was confirmed. B, concentration of intracellular putrescine in the different strains.
Comparison of Kinetic Parameters of YeeF with Previously Reported Importers
E. coli has four putrescine importers: PotABCD (30), PotE (18), PotFGHI (15), and PuuP (20). The kinetic values of putrescine uptake by these importers were determined using bacteria harboring pACYC184 with potABCD, potE, potFGHI, or puuP. To compare the kinetic parameters of YeeF with those of putrescine importers reported in previous studies, we constructed a pACYC184-yeeF+/ΔpotABCD ΔpotE ΔpotFGHI ΔpuuP ΔydcSTUV ΔyeeF strain (SK629) and measured the Km and Vmax values of putrescine uptake by YeeF in SK629 cells grown in LBG (supplemental Fig. S1 and Table 2). Surprisingly, the Km of YeeF (155 μm) was 40 to 300 times higher than that of other importers previously reported (Table 2). On the other hand, the Vmax of YeeF (9.3 nmol/min/mg) was comparable to that of PotABCD, PotFGHI, and PuuP.
TABLE 2.
Kinetic parameters of the polyamine importers
YeeF Is a Major Putrescine Importer in LBG-grown E. coli
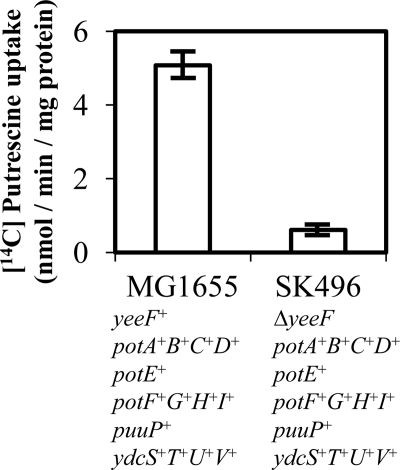
E. coli has several putrescine importers. It seems that E. coli selects one or more putrescine importers by switching gene expression according to growth environment. To show that YeeF is the main putrescine importer of E. coli cells grown in LBG, we constructed a ΔyeeF single knock-out mutant (SK496) and performed a transport assay using 500 μm [14C]putrescine, a concentration that is more than three times higher than the Km value of YeeF. The putrescine uptake activity of SK496 (ΔyeeF potA+B+C+D+ potE+ potF+G+H+I+ puuP+ ydcS+T+U+V+) was reduced by 88% as compared with MG1655 (yeeF+ potA+B+C+D+ potE+ potF+G+H+I+ puuP+ ydcS+T+U+V+) (Fig. 5). This result clearly demonstrated that YeeF is the main putrescine importer in E. coli grown in LBG when the amount of putrescine in the environment is in the order of hundreds of μm.
FIGURE 5.
YeeF is a major putrescine importer in E. coli grown in LBG medium. The ΔyeeF single knock-out mutant shows an impaired putrescine uptake. The strains MG1655 and SK496 were inoculated in 100-ml LBG medium in 500-ml Erlenmeyer flasks. The initial optical density of the culture, measured at 600 nm, was adjusted to 0.001. The flasks were shaken at 150 rpm at 37 °C. Bacterial cells were harvested at A600 = 1.2–1.3 and washed once with cold M9-glucose. The cells were incubated with 500 μm [14C]putrescine, and the reaction was terminated after 2, 4, 6, 8, or 10 min. The linearity between the uptake of [14C]putrescine and the incubation time was confirmed. The assays were performed three times with independent bacterial cultures. Values are expressed as the mean ± S.D.
Surface Motility Induced by YeeF and Putrescine Was Type 1 Pili-driven
It is well known that the flagella-driven swarming motility of E. coli contributes to its surface motility. However, the surface motility induced by YeeF and putrescine observed in the present study was not influenced by the deletion of fliC encoding the basic subunit of a flagella filament (Fig. 6A). In addition to the flagella, there are pili on the cell surface of E. coli. The deletion of fimA, which encodes the major subunit of E. coli type 1 pili, completely abolished the surface motility induced by YeeF and putrescine (Fig. 6A). In addition, in the wild-type genetic background, the deletion of fliC did not influence the surface motility; in contrast, the ΔfimA strain completely lost all surface motility (Fig. 6B). In the complementation experiment (Fig. 6C), both the fimA+ strain (SK653) and the ΔfimA strain complemented with a plasmid vector containing the fimA+ gene (SK655) exhibited surface motility on LBGS plates supplemented with 400 μm putrescine, but not on putrescine-free LBGS plates. In contrast, the ΔfimA strain (SK654) did not exhibit surface motility on LBGS plates, whether supplemented with 400 μm putrescine or not. These results showed that the surface motility induced by YeeF-imported putrescine observed in the present study was not mainly driven by flagella but instead by type-1 pili.
FIGURE 6.
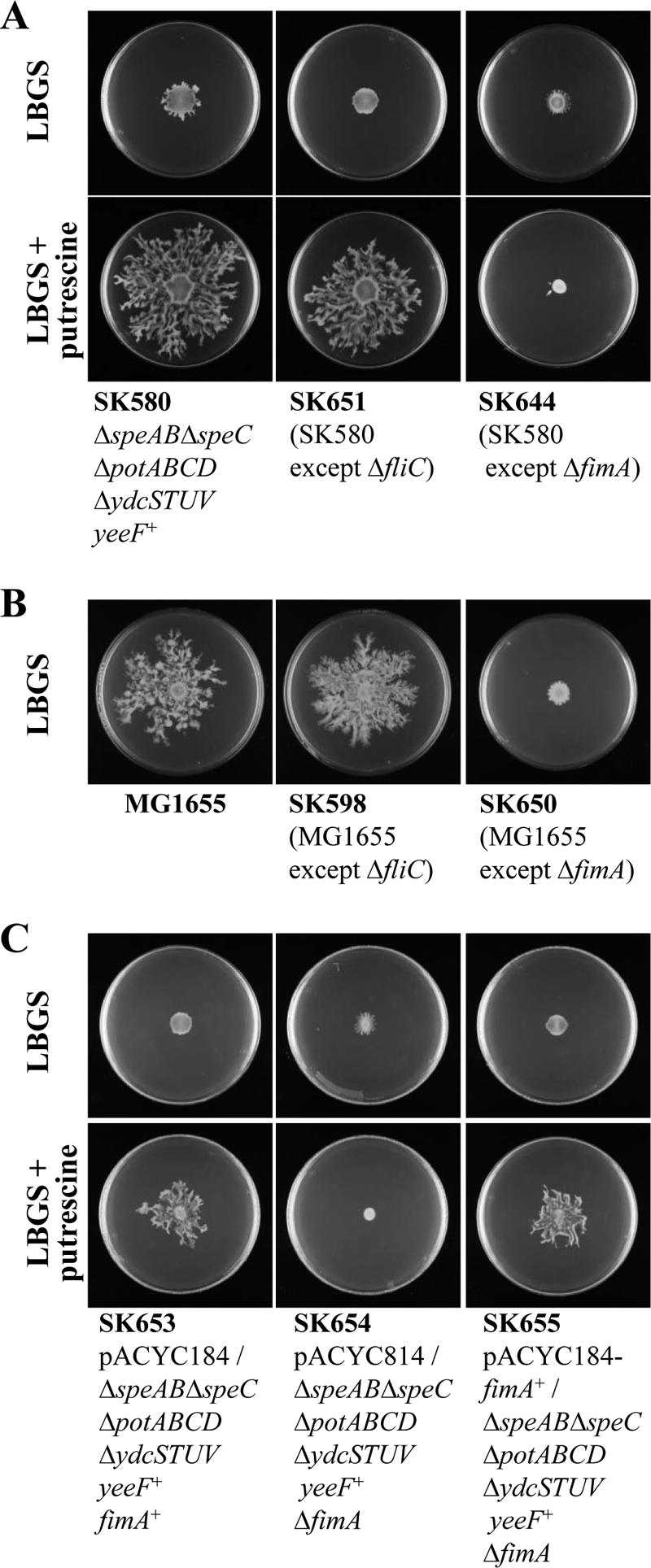
fimA, not fliC, is indispensable in the surface motility observed in the present study. Bacterial cultures were diluted in LB medium to an A600 of 0.4, and 3 μl of the diluted culture were spotted onto the center of LBGS plates, supplemented with or without 400 μm putrescine, and incubated at 37 °C. The plates were photographed 40 h after inoculation. In C, the medium was supplemented with chloramphenicol (15 μg/ml) to maintain the plasmids. A, the surface motility induced by putrescine and yeeF is fimA-dependent, not fliC-dependent. B, the surface motility depended on fimA, not fliC, of the strains in the wild-type background. C, complementation experiment with fimA.
DISCUSSION
Proposal of a New Gene Name for yeeF
YeeF is a novel major putrescine importer (Figs. 4 and 5B) required for extracellular putrescine-induced surface motility (Fig. 3). YeeF has low affinity for putrescine (supplemental Fig. S1 and Table 2). On the basis of this characteristic, we propose the new name PlaP (putrescine low affinity permease).
Surface Motility Observed in the Present Study
In the present study, E. coli exhibited active surface motility on semisolid agar containing 0.6% Eiken agar, which is widely used for studying swarming in E. coli. It is well known that the swarming motility is driven by the flagella, and FliC is the basic subunit that polymerizes to form the rigid flagellar filament of E. coli. In the present study, however, the ΔfliC strain exhibited the same active surface motility as the fliC+ strain (Fig. 6). On the other hand, the deletion of fimA, which encodes the basic component of type 1 pili, did not exhibit any surface motility. These results indicate that the surface motility observed in the present research is driven mainly by type 1 pili. Nonetheless, it is possible that surface motility observed in the present study is partially driven by flagella because the synthetic pathway of flagellum was significantly up-regulated (supplemental Table S1). Twitching was previously described as a type of surface motility driven by type 4 pili, but the fact that E. coli exhibits surface motility driven by type 1 pili is a new discovery. In the previous report (31), flagellum-independent surface motility was also observed in E. coli, and it is highly likely that the surface motility observed in the previous report is type 1 pili-driven.
In another previous report (28), the surface motility of JW1908 (ME9062 except ΔfliC) was abolished. Accordingly, we confirmed that JW1908 exhibits a defect in surface motility. On the contrary, SK598 (MG1655 except ΔfliC), like MG1655, exhibited surface motility (Fig. 6B). This contradiction probably arises from the genetic differences between the parental strains of JW1908 and SK598 (see Table 1).
Relationship between Surface Motility Driven by fimA and Intracellular Putrescine
In E. coli cells, polyamines stimulate the synthesis of many types of proteins at the level of translation without a change in the transcription level. The genes that encode proteins regulated by this mechanism are referred to as the “polyamine modulon” (8). The translation of mRNAs encoded by the genes classified into polyamine modulon is inefficient without polyamine because the Shine-Dalgarno sequence is more distant from the initiation codon AUG compared with normal mRNAs. Alternatively, this could be explained by the fact that the initiation codon of mRNA is UUG or GUG, which is less effective than AUG, or that the mRNA has a termination codon in the middle of the ORF. Polyamine induces a conformational change in those types of mRNAs and enhances their translation efficiency. SK595 does not have intracellular putrescine and does not exhibit any surface motility; in contrast, SK591 and SK594 have intracellular putrescine imported by PlaP and exhibit surface motility (Figs. 3 and 4). The transcription of fimA was not up-regulated between SK591 and SK595 or between SK594 and SK595 (data not shown). Therefore, it was possible that fimA was regulated in the same manner as genes classified into the polyamine modulon; however, fimA mRNA does not have the above-mentioned characteristics and it is not thought that the level of FimA is enhanced by putrescine at the level of translation. A genome-wide gene expression comparison revealed that many nutrient metabolic pathways were significantly up-regulated in SK591 and SK594, which exhibit surface motility, compared with SK595, which does not exhibit surface motility (supplemental Tables S1 and S2). Therefore, putrescine imported by PlaP does not stimulate either the transcription or translation of fimA but the energy produced by the nutrient metabolic pathways up-regulated by putrescine may contribute to the motion of FimA and thereby to active surface motility.
Significance of Multiple Importers of E. coli
In previous studies, four putrescine importers have been reported in E. coli: PotABCD (16, 17), PotFGHI (15), PotE (18), and PuuP (20). In the present report, PlaP, a novel putrescine importer, was identified. The number of putrescine importers has thus increased to five.
In the present study, the deletion of genes encoding other putrescine importers, with the exception of ΔplaP, had no influence on the surface motility on LBGS plates (Fig. 2). In addition, SK595 cells grown in LBG supplemented with 400 μm putrescine could not take up putrescine, even though their genotype is potE+ potF+G+H+I+ puuP+ (Fig. 4). Furthermore, the putrescine uptake activity of the ΔplaP single knock-out mutant (potE+ potF+G+H+I+ puuP+ ydcS+T+U+V+) grown in LBG medium was reduced by 88% as compared with the parental strain MG1655 (Fig. 5). These results indicated that potE, potFGHI, and puuP were not expressed under the culture conditions used in the present study.
The growth conditions of E. coli in this study are thought to down-regulate the expression of genes encoding other putrescine importers. For example, the M9-tryptone used in the PuuP study does not contain glucose (20), whereas LBGS and LBG contain 0.5% glucose. It is thought that the gene expression of the five (or more) putrescine importers in E. coli changes depending on the environment.
Significance of a Low-affinity Putrescine Importer
The concentration of putrescine in the human intestinal tract, which is one of the environments that E. coli inhabits, is in the order of tens to hundreds of μm (32). Therefore, the Km value of PlaP, 154 μm is not improbable. However, compared with other putrescine importers previously reported, the affinity of PlaP for putrescine is remarkably low (Table 2). Because putrescine concentrations in the medium acted as a trigger for surface motility of E. coli harboring plaP+ (Figs. 1–3) the low-affinity putrescine importer PlaP must be of physiological significance.
In a process termed quorum sensing, bacteria produce, release, and respond to signaling molecules, which accumulate as a function of cell density. It was previously reported that the amount of putrescine in culture exported by E. coli increased roughly in proportion to the degree of growth (33). Also, we observed that the concentration of putrescine in culture exported by E. coli reached 180 μm when strain SH639 cultured in M9-tryptone medium (26) under low aeration condition. If E. coli uses putrescine as an extracellular signal to sense cell density as in the quorum sensing signaling system, the concentration of putrescine must be kept at levels to reflect the cell density. The low affinity of PlaP for putrescine (Table 2) may allow E. coli to sense the cell density depending on the concentration of extracellular putrescine because PlaP cannot decrease extracellular putrescine effectively when the concentration of putrescine is below the Km value of PlaP.
The PlaP homolog is found in Y. pestis and P. mirabilis where putrescine has been reported previously to function as a signaling molecule (11, 12). The low affinity transporter PlaP may be widely used in the bacterial world to conduct putrescine-regulated cell-to-cell communication.
Potential Application of Observations Reported in Present Study
The present study may allow us to interrupt the cell-to-cell communication of the above-mentioned pathogenic bacteria and to attenuate their pathogenicity by inhibiting uptake of putrescine by PlaP. This inhibition can be performed by either a PlaP inhibitor or by plaP repression. It was previously reported that plaP transcription is repressed by acetate (34) or sodium salicylate (35); therefore, it is suspected that the plaP gene is regulated by many factors. It may not be difficult to attenuate the pathogenicity of bacteria by repressing plaP transcription.
Acknowledgments
We thank the National BioResource Project E. coli (National Institute of Genetics, Japan), for providing strains ME9062, JW1908, and JW4277.
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry by the Bio-oriented Technology Research Advancement Institution. A part of this work was supported by Grant-in-aid for Scientific Research 21380059 from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Fig. S1.
- LBGS
- LB glucose semisolid.
REFERENCES
- 1. Kuehn G. D., Rodriguez-Garay B., Bagga S., Phillips G. C. (1990) Plant Physiol. 94, 855–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Terui Y., Ohnuma M., Hiraga K., Kawashima E., Oshima T. (2005) Biochem. J. 388, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tabor C. W., Tabor H. (1985) Microbiol. Rev. 49, 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pegg A. E. (1986) Biochem. J. 234, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerner E. W., Meyskens F. L., Jr. (2004) Nat. Rev. Cancer 4, 781–792 [DOI] [PubMed] [Google Scholar]
- 6. Igarashi K., Kashiwagi K. (2000) Biochem. Biophys. Res. Commun. 271, 559–564 [DOI] [PubMed] [Google Scholar]
- 7. van Dam L., Korolev N., Nordenskiöld L. (2002) Nucleic Acids Res. 30, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Igarashi K., Kashiwagi K. (2006) J. Biochem. 139, 11–16 [DOI] [PubMed] [Google Scholar]
- 9. Boyle S. M., Markham G. D., Hafner E. W., Wright J. M., Tabor H., Tabor C. W. (1984) Gene 30, 129–136 [DOI] [PubMed] [Google Scholar]
- 10. Tabor C. W., Tabor H., Xie Q. W. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6040–6044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel C. N., Wortham B. W., Lines J. L., Fetherston J. D., Perry R. D., Oliveira M. A. (2006) J. Bacteriol. 188, 2355–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sturgill G., Rather P. N. (2004) Mol. Microbiol. 51, 437–446 [DOI] [PubMed] [Google Scholar]
- 13. Karatan E., Duncan T. R., Watnick P. I. (2005) J. Bacteriol. 187, 7434–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurihara S., Suzuki H., Tsuboi Y., Benno Y. (2009) FEMS Microbiol. Lett. 294, 97–101 [DOI] [PubMed] [Google Scholar]
- 15. Pistocchi R., Kashiwagi K., Miyamoto S., Nukui E., Sadakata Y., Kobayashi H., Igarashi K. (1993) J. Biol. Chem. 268, 146–152 [PubMed] [Google Scholar]
- 16. Furuchi T., Kashiwagi K., Kobayashi H., Igarashi K. (1991) J. Biol. Chem. 266, 20928–20933 [PubMed] [Google Scholar]
- 17. Kashiwagi K., Hosokawa N., Furuchi T., Kobayashi H., Sasakawa C., Yoshikawa M., Igarashi K. (1990) J. Biol. Chem. 265, 20893–20897 [PubMed] [Google Scholar]
- 18. Kashiwagi K., Miyamoto S., Suzuki F., Kobayashi H., Igarashi K. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4529–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kurihara S., Oda S., Kato K., Kim H. G., Koyanagi T., Kumagai H., Suzuki H. (2005) J. Biol. Chem. 280, 4602–4608 [DOI] [PubMed] [Google Scholar]
- 20. Kurihara S., Tsuboi Y., Oda S., Kim H. G., Kumagai H., Suzuki H. (2009) J. Bacteriol. 191, 2776–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller J. H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, pp. 263–274 and 437, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 23. Kim S. Y., Volsky D. J. (2005) BMC Bioinformatics 6, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kato J., Hashimoto M. (2007) Mol. Syst. Biol. 3, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koyanagi T., Katayama T., Suzuki H., Kumagai H. (2004) J. Bacteriol. 186, 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kurihara S., Oda S., Tsuboi Y., Kim H. G., Oshida M., Kumagai H., Suzuki H. (2008) J. Biol. Chem. 283, 19981–19990 [DOI] [PubMed] [Google Scholar]
- 27. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 28. Inoue T., Shingaki R., Hirose S., Waki K., Mori H., Fukui K. (2007) J. Bacteriol. 189, 950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyamoto S., Kashiwagi K., Ito K., Watanabe S., Igarashi K. (1993) Arch. Biochem. Biophys. 300, 63–68 [DOI] [PubMed] [Google Scholar]
- 30. Kashiwagi K., Endo H., Kobayashi H., Takio K., Igarashi K. (1995) J. Biol. Chem. 270, 25377–25382 [DOI] [PubMed] [Google Scholar]
- 31. Brown II, Häse C. C. (2001) J. Bacteriol. 183, 3784–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matsumoto M., Kakizoe K., Benno Y. (2007) Microbiol. Immunol. 51, 37–46 [DOI] [PubMed] [Google Scholar]
- 33. Schiller D., Kruse D., Kneifel H., Krämer R., Burkovski A. (2000) J. Bacteriol. 182, 6247–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnold C. N., McElhanon J., Lee A., Leonhart R., Siegele D. A. (2001) J. Bacteriol. 183, 2178–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pomposiello P. J., Bennik M. H., Demple B. (2001) J. Bacteriol. 183, 3890–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suzuki H., Koyanagi T., Izuka S., Onishi A., Kumagai H. (2005) J. Bacteriol. 187, 5861–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suzuki H., Kumagai H., Tochikura T. (1987) J. Bacteriol. 169, 3926–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]