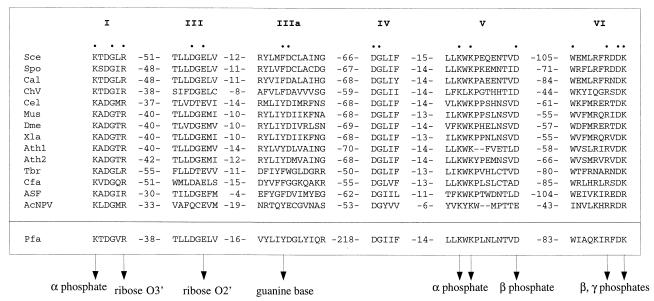
Figure 2.
Guanylyltransferase signature motifs present in metazoan, plant, and viral capping enzymes are conserved in P. falciparum Pgt1. The amino acid sequences are aligned for the capping enzymes of S. cerevisiae (Sce), S. pombe (Spo), C. albicans (Cal), Chlorella virus PBCV-1 (ChV), Caenorhabditis elegans (Cel), mouse (Mus), Drosophila melanogaster (Dme), Xenopus laevis (Xle), Arabidopsis thaliana (Ath), Trypanosoma brucei gambiense (Tbr), Crithidia fasciculata (Cfa), African swine fever virus (ASF), and AcNPV baculovirus (AcNPV). The motifs of the P. falciparum (Pfa) guanylyltransferase are highlighted below the other aligned sequences. The numbers of amino acid residues separating the motifs are indicated. The amino acids of the Sce guanylyltransferase that are essential for function in vivo are denoted by dots. Specific contacts between amino acid side chains and the GTP substrate in the ChV capping enzyme–GTP cocrystal are indicated by arrowheads.