Abstract
RNase H2 cleaves RNA sequences that are part of RNA/DNA hybrids or that are incorporated into DNA, thus, preventing genomic instability and the accumulation of aberrant nucleic acid, which in humans induces Aicardi-Goutières syndrome, a severe autoimmune disorder. The 3.1 Å crystal structure of human RNase H2 presented here allowed us to map the positions of all 29 mutations found in Aicardi-Goutières syndrome patients, several of which were not visible in the previously reported mouse RNase H2. We propose the possible effects of these mutations on the protein stability and function. Bacterial and eukaryotic RNases H2 differ in composition and substrate specificity. Bacterial RNases H2 are monomeric proteins and homologs of the eukaryotic RNases H2 catalytic subunit, which in addition possesses two accessory proteins. The eukaryotic RNase H2 heterotrimeric complex recognizes RNA/DNA hybrids and (5′)RNA-DNA(3′)/DNA junction hybrids as substrates with similar efficiency, whereas bacterial RNases H2 are highly specialized in the recognition of the (5′)RNA-DNA(3′) junction and very poorly cleave RNA/DNA hybrids in the presence of Mg2+ ions. Using the crystal structure of the Thermotoga maritima RNase H2-substrate complex, we modeled the human RNase H2-substrate complex and verified the model by mutational analysis. Our model indicates that the difference in substrate preference stems from the different position of the crucial tyrosine residue involved in substrate binding and recognition.
Keywords: DNA Repair, Protein-DNA Interaction, Protein Structure, Ribonuclease, X-ray Crystallography, Aicardi-Goutières Syndrome, RNA/DNA Hybrid, RNase H2
Introduction
Ribonucleases H (RNases H) are nucleases that cleave RNA/DNA hybrids (1). The two main groups of RNases H (type 1 or RNases H1 and type 2 or RNases H2) share low sequence similarity and differ in their biochemical properties (2). However, type 1 and type 2 RNases H have the same fold of the catalytic core, called the RNase H fold, a common feature of other related enzymes, such as integrases, transposases, Argonaute, and RuvC resolvase (3). They all belong to the retroviral integrase superfamily. The key feature of the RNase H fold is a β-sheet of three anti-parallel and two parallel strands. The active site of retroviral integrase superfamily proteins is predominantly composed of carboxylates that coordinate two divalent metal ions that are essential for catalysis. The preferred metal ions are Mg2+, but Mn2+ can also support the reaction. Type 1 enzymes require the presence of at least four ribonucleotides and hydrolyze the nucleic acid in the middle of the RNA sequence (2). The most unique and possibly most important feature of type 2 enzymes is their ability to cleave preferentially on the 5′ side of the last ribonucleotide in DNA-RNA-DNA/DNA substrates. RNases H2 hydrolyze these substrates even when a single ribonucleotide is present. Single ribonucleotides can result from misincorporation by DNA polymerases, which occurs frequently in eukaryotic cells (4). The presence of these ribonucleotides in the DNA can lead to genomic instability, and they must be removed (5–7). RNase H2 works together with FEN-1 nuclease to completely excise single ribonucleotides from the DNA (5, 6).
RNases H2 are present in all kingdoms of life. Bacterial enzymes contain a single subunit and prefer substrates in which a ribonucleotide has its 3′-phosphate connected to a deoxynucleotide (-XpRpDp-) being indifferent to the type of nucleotide on its 5′ side; RNA primers of Okazaki fragments and single ribonucleotides in duplex DNA are such substrates. The products after digestion of these substrates are a single ribonucleotide on the 5′-end of the downstream DNA (pRpDp-). Eukaryotic proteins consist of a catalytic subunit (RNase H2A), with striking similarities to the prokaryotic enzymes, and two auxiliary subunits (RNase H2B and RNase H2C) (8). The latter share no significant sequence similarity with known proteins. Their roles are unclear, but they are required for activity (8, 9). Eukaryotic RNases H2 cleave the same substrates as their bacterial homologs but also cleave an RNA/DNA hybrid in which no -RpDp- junction exists. Moreover, eukaryotic RNases H2 cleave RNA/DNA hybrids processively, whereas the prokaryotic enzymes act in a distributive manner. Archaeal RNases H2 have substrate preferences similar to the eukaryotic enzymes.
Human type 2 RNase H is the main RNase H activity in human cells (10, 11). Mutations in any of the three subunits of the human enzyme can result in Aicardi-Goutières syndrome (AGS)2 (12, 13), an autosomal recessive genetic disorder with symptoms similar to in utero viral infection that severely affects the nervous system. Inactivation of the enzyme can lead to the accumulation of RNA/DNA hybrids that in turn activates the innate immune response, leading to an infection-like phenotype (12). Because no AGS patient has been observed with a complete loss of RNase H2, type 2 RNase H activity has been suggested to be essential in humans (12), although Saccharomyces cerevisiae does not require RNase H1 or RNase H2 (7).
Crystal structures of archaeal and bacterial type 2 RNases H are available (14–16). In addition to the catalytic domain adopting the RNase H fold, they also contain a helical C-terminal domain. We recently reported the first crystal structures of bacterial RNase H2 in complex with nucleic acid, a 12-mer double-stranded DNA with a single ribonucleotide embedded in one of the strands (17). They demonstrated that the substrate is bound in a cleft between the catalytic and C-terminal domains. The 5′ phosphate of the (5′)RNA-DNA(3′) junction is located at the active site, and the 2′-OH group of the ribonucleotide interacts with conserved glycine, arginine, and glycine (GRG motif). An absolutely conserved tyrosine residue from the C-terminal domain forms a hydrogen bond with this 2′-OH group and a stacking interaction with the second residue of the junction. This stacking is the most efficient if no 2′-OH group is present in the ribose ring and, hence, selects for DNA, which leads to specific binding of the RNA-DNA junction. The stacking interaction with tyrosine also introduces a deformation of the substrate, allowing the phosphate group in the middle of the junction to participate in the coordination of a Mg2+ ion at the active site. Such substrate preference is lost in the presence of a Mn2+ ion because Mn2+ binding is not coupled to substrate deformation.
While we were working on the determination of the structure of the human RNase H2 complex, the first structure of a eukaryotic RNase H2 from mouse was reported (18) showing that the catalytic subunit of the complex closely resembles the known structures of RNases H2 and that the auxiliary subunits form a highly intertwined dimer adopting a triple-barrel fold. Our human structure was solved at 3.1 Å resolution, and it differed from the mouse structure in tracing of the B and C subunits. In our refined structure, we have been able to map the positions of all currently reported RNase H2 mutations in AGS patients. Because of the similarities between the catalytic subunits of human and mouse RNases H2A and the monomeric Thermotoga maritima RNase H2, we used our bacterial RNase H2 complex structure (17) to build a model of substrate binding by the human enzyme, which we verified through mutagenesis studies.
EXPERIMENTAL PROCEDURES
Protein Preparation
To allow the testing of different combinations of truncated subunits, subunit A was cloned into a pET28 expression vector, and subunits B and C were cloned into a pET15 vector (EMD Biochemicals). All proteins carried N-terminal His tags removable with PreScission Protease (subunit A) or thrombin (subunits B and C). The mutagenesis of the constructs was performed using the QuikChange kit (Stratagene) or inside-out PCR.
pET28-A and pET15-BC vectors with appropriate deletions were co-transformed into Escherichia coli BL21 cells for co-expression. Protein expression was induced overnight with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside at 30 °C. Bacterial cells were next suspended in 40 mm NaH2PO4 (pH 7.0), 100 mm NaCl, and 5% glycerol with the addition of a mixture of protease inhibitors and incubated on ice in the presence of 1 mg/ml lysozyme. After sonication, the cleared lysate was applied to a HisTrap column (GE Healthcare) equilibrated with 10 mm imidazole, 40 mm NaH2PO4, 0.5 m NaCl, and 5% glycerol. After a wash step with 60 mm imidazole, the protein was eluted with a linear gradient of imidazole from 60 to 300 mm. Fractions containing the protein were dialyzed overnight against 40 mm NaH2PO4 (pH 7.0), 100 mm NaCl, 5% glycerol, 1 mm DTT, and 0.5 mm EDTA. The His tags were removed by overnight incubation with thrombin and PreScission Protease. The protease-digested sample was applied to a heparin column (GE Healthcare) equilibrated with 40 mm NaH2PO4 (pH 7.0), 0.1 m NaCl, 5% glycerol, 1 mm DTT, and 0.5 mm EDTA, and the protein was eluted with a linear gradient of NaCl from 0.1 to 0.5 m. Selected fractions were next concentrated and applied to a gel filtration column (Superdex 200, GE Healthcare). The final protein sample was concentrated to 35–45 mg/ml and stored in 20 mm HEPES (pH 7.0), 75 mm NaCl, 5% glycerol, 1 mm DTT, and 0.5 mm EDTA.
Crystallization and Structure Determination
We previously constructed an E. coli system to overproduce a human RNase H2 mutant with the PCNA-interacting motif truncated (ABΔPIPC) (9). The initial crystallization trials with ABΔPIPC yielded crystals in a form of very thin and flexible plates or spherulites that could not be improved through optimization of the crystallization conditions. Deletion mutant AB14–233C crystallized more readily than the ABΔPIPC and yielded larger and regular crystals. For crystallization, the protein solution (13 mg/ml) was mixed with the reservoir solution at equal volumes and crystallized by the sitting drop vapor diffusion method at 18 °C. The crystals of AB14–233C complex were obtained with 0.1 m MgCl2, 15% PEG 3350, 0.1 m Bis-Tris (pH 5.5), and 2 mm reduced glutathione. The crystals were transferred stepwise to dehydrating solutions with increasing PEG 3350 concentrations up to a final concentration of 40% and flash-frozen in liquid nitrogen.
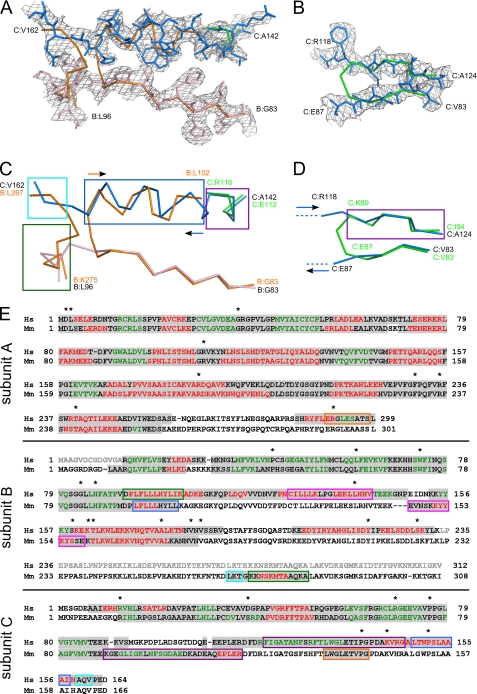
The diffraction data of the AB14–233C complex crystals were collected at the European Synchrotron Radiation Facility at beamline 23-2 on a Mar225 CCD detector at 100 K. The dataset was processed and scaled using HKL2000 (19). The crystals belong to the P1 space group and contain six RNase H2 complexes per asymmetric unit. The structure was solved by the molecular replacement method using the mouse RNase H2 structure (Protein Data Bank ID 3KIO) and Phaser program (20). The resulting model was refined using phenix.refine (21) and autoBuster (Global Phasing Ltd, Cambridge, UK). After several rounds of refinement it was apparent that the model from the molecular replacement did not fit well to electron density maps. Rebuilding the dubious regions involved changes to the previously assumed tracing and chain assignment. The most significant changes were introduced to the region defined in the human protein by two segments, B:83–96 and C:142–162 (Fig. 2, A, C, and E). Starting from residue B:Gly-83, the B chains of both human and mouse protein superimpose well until B:Val-91 (B:Met-91 in mouse RNase H2). Here, the B chain of the human protein continues in the same direction, whereas the mouse B chain makes a sharp turn and forms an α-helix. In the human structure this region is also a helix but belongs to subunit C (residues 148–158) and runs in the opposite direction. The other end of this helix is built in the mouse structure as a fragment of the C subunit (residues 112–116), which also adopts a helical structure, this time with the same polarity as the human C chain. Therefore, the single helix in the human structure (C:142–158) in the mouse structure is built as two helices coming from different chains and with opposite polarity. The mouse structure also contains a stray B:267–275 strand built into electron density, which we assigned to two fragments of the human protein: B:92–96 and C:158–161. The need to rebuild this region was justified by both the observation that the electron density in this area is clearly split in two traces and the fact that the deletion mutant of the B subunit we used for crystallization was C-terminally truncated at residue 233 and, thus, lacked the fragment corresponding to mouse residues B:267–275.
FIGURE 2.
Tracing differences between the human and mouse RNase H2 structures. A and B, two regions with the largest differences in tracing between human and mouse structures are shown. The human structure is shown in stick representation and colored in light pink for subunit B and blue for subunit C. The backbone trace of the mouse structure is shown in orange (subunit B) and green (subunit C). A simulated annealing omit electron density map from the human structure contoured at 1σ is overlaid on the superimposition. C and D, the same views are shown but with both proteins shown as a backbone trace. In C, the polarities of the helices are shown with arrows colored as the protein subunits. E, sequence alignment of human (Hs) and mouse (Mm) RNases H2. The sequence highlighted in gray is observed in respective structures. The sequence colored in red corresponds to α-helices, and the sequence colored in green corresponds to β-strands. The sequence shown in gray lettering was removed from the human complex construct used for crystallization. The regions that have different tracing between human and mouse models are boxed. Corresponding colors denote sequences that are built in the same locations in the human and mouse structures. These regions are also indicated in C and D using boxes of the same color. Asterisks show the positions of AGS mutations.
Another fragment that was rebuilt in the human structure is the region corresponding to the mouse residues C:83–94 (Fig. 2, B, D, and E). Whereas the C:83–87 region forming a β-strand superimposes well with the human complex model, the C:89–94 part appears to fall out of register. The lack of density bridging the two β-strands in this region together with the analysis of the downstream sequence led us to assign residues C:119–124 in place of mouse C:89–94. Thus, instead of taking a sharp turn built in the mouse structure, the C chain of the human structure continues beyond that turning point into a non-visible and, hence, likely unstructured stretch of ∼30 residues and then resumes as the antiparallel β-strand.
The structures of the A subunits of the mouse and human proteins are much more consistent. They are also similar to the published archaeal and bacterial structures, with the exception of the C-terminal fragment of the A subunit (∼50 residues), which is only present in eukaryotic RNases H2. A substantial proportion of this region is not visible in the mouse and human RNase H2 structures. However, because of the corrections we made to the C chain, we were able to trace the very last 16 residues of the A subunit that were misassigned to the C chain in the mouse model (Fig. 2E).
The final rebuilt model was refined in autoBuster interspersed with manual building in Coot (22). Structural analyses, including superpositions and structural figures, were prepared in PyMol (Schrödinger). The coordinates of the structure have been deposited in the Protein Data Bank under the accession code 3PUF.
RNase H Activity Assays
RNase H activity was determined using a uniformly [α-32P]ATP-labeled poly(rA)/poly(dT) substrate by measuring the amount of radioactivity of the acid-soluble digestion product in various solution conditions as described previously (23). For assays with Mg2+, the enzyme concentrations were 0.02 and 0.2 nm, and for assays in the presence of Mn2+, the enzyme concentrations were 0.2 and 4 nm. For short substrate experiments, the 5′-32P-labeled 12-mer substrates (the sequences of the oligonucleotides are given in supplemental Fig. 3) were digested with native and mutant proteins in 15 mm Tris-HCl (pH 7.9), 50 mm NaCl, 1 mm DTT, 100 μg/ml BSA, 5% glycerol, and 10 mm MgCl2 (or 1 mm MnCl2). The hydrolysis products were analyzed on 20% Tris borate EDTA-urea polyacrylamide gels. The reaction products were visualized by phosphorimaging.
Thermofluor Assay
The variants of human RNase H2 AB14–233C B:W73L, B:G83S, B:H86R, and B:Y219H were purified on a nickel column essentially using the same protocol as for wild type AB14–233C. The thermal stability of these proteins was analyzed using a ThermoFluor assay according to the published protocol with a few modifications (24). Proteins at 2 μm concentration were prepared in a 40-μl volume in 50 mm HEPES (pH 7.2), 100 mm NaCl, and 5× SYPRO Orange (Invitrogen). Thermal denaturation of the proteins was monitored by measuring the fluorescence intensity using a SYBR Green filter on LightCycler480 (Roche Applied Science). The temperature of the protein solutions was linearly increased by 0.03 °C/s from 20 to 90 °C. The melting temperature (Tm) was defined as the temperature showing the maximum fluorescence gradient.
RESULTS AND DISCUSSION
Complex Formation and Structure Determination of Human RNase H2
Eukaryotic RNases H2 are considerably more complex enzymes than their prokaryotic homologs. The catalytic subunit (A) has N- and C-terminal extensions that are not present in the prokaryotic enzymes. The B and C accessory subunits are unique to eukaryotic RNases H2, and although their functions are unknown, they are assumed to assist the catalytic subunit. To determine which regions of the different subunits are important for complex formation, we made several deletions and tested the solubility and activity of the mutated complexes and individual subunits (supplemental Table 1). Deletion of the N- and C-terminal ends of the B subunit (a protein comprising residues 14–233) formed a complex that retained close to wild type activity and solubility. However, deletion of the N- (A24–299) or C-terminal (A1–250) extensions of the A subunit produced insoluble protein that could not form a complex with the B and C subunits when they were co-expressed in E. coli. In our hands the expression of RNase H2A alone resulted in mostly insoluble protein, and the small amount of enzyme that could be partially purified in soluble form was inactive for the cleavage of RNA/DNA and RNA-DNA/DNA hybrid substrates (data not shown).
We obtained crystals of the human RNase H2 AB14–233C that diffracted x-rays to 3.1 Å resolution. We used the recently published structure of mouse RNase H2 (18) (Protein Data Bank ID 3KIO) as a search model in molecular replacement to solve the human RNase H2 structure. After several rounds of refinement, although the overall structures of the mouse and human complexes were very similar, some regions of the model needed to be rebuilt. This particularly involved the middle portions of the B and C subunits (see “Experimental Procedures”).
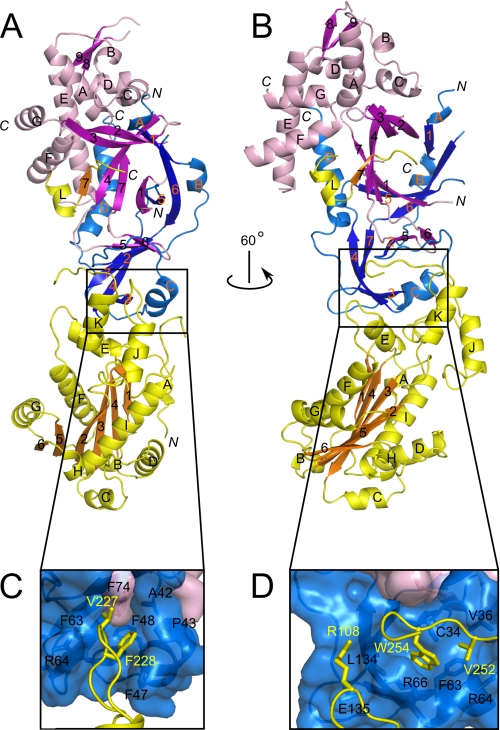
The human structure was refined to a final R factor of 20.6% and Rfree of 25.5% (Table 1). The asymmetric unit of our crystals accommodates 6 copies of the human RNase H2 AB14–233C complex. Supplemental Table 2 provides the ranges of the residues that were traced in each copy of the complex. The six complexes were refined independently (non-crystallographic symmetry was not applied) and are essentially identical; they can be superimposed with a 0.74 Å root mean square deviation of the positions of 508 Cα atoms from each copy of the complex (supplemental Fig. 1). The three subunits of the complex are arranged in one line, with the C subunit located in the middle and B and A subunits on its sides (Fig. 1, A and B). The B and C subunits form an intertwined dimer that adopts a triple-barrel fold with a central core of 14 β-strands. The catalytic A subunit interacts almost exclusively with subunit C. An α-helix (residues 98–108) and three loops (residues 196–199, 227–232, and 250–256) of the A subunit make mostly hydrophobic contacts with four regions of the C subunit (residues 34–36, 47–51, 62–67, and 134–135) (Fig. 1, C and D). The extreme C-terminal fragment of subunit A (residues 284–299) also forms extensive contacts with subunits B and C, and residues 292–296 add a short β-strand to the central β-sheet of the triple-barrel of subunits B and C. Deletion of one of the regions described above (AΔ104–110) prevented complex formation (supplemental Table 1). We hypothesized that removing the hydrophobic regions of the catalytic subunit that are necessary for complex formation might yield a soluble and active single subunit RNase H2. To accomplish this, we deleted the hydrophobic region involved in complex interactions shown in Fig. 1D and C-terminal extension to produce A1–250,Δ104–110. Although the solubility of the mutated protein was higher than the wild type 2A, the protein was inactive (data not shown). To completely remove the regions of interactions with the accessory subunits, we also replaced the hydrophobic residues of the A subunit shown in Fig. 1C with the corresponding region of T. maritima RNase H2, which is soluble and active as a monomer. The resulting protein (A1–250,Δ104–110,Δ223–231,NGVL) remained inactive (data not shown), demonstrating the importance of the complex formation for activity. The complete description of the differences in tracing between mouse and human RNases H2 is given under “Experimental Procedures” and shown in Fig. 2.
TABLE 1.
Crystallographic data collection and refinement statistics for human RNase H2 structure
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 90.1, 108.4, 114.3 |
| α, β, γ (°) | 105.9, 103.7, 111.4 |
| Wavelength (Å) | 0.87260 |
| Resolution (Å)a | 50-3.1 (3.15-3.10) |
| Rmerge | 10.7 (41.1) |
| I/σ(I) | 11.8 (2.2) |
| Completeness (%) | 97.1 (87.6) |
| Redundancy | 2.8 (2.3) |
| Refinement | |
| Resolution (Å) | 3.1 |
| No. reflections | 63130 |
| Rwork/Rfree (%) | 20.6/25.5 |
| No. atoms | 25,820 |
| Protein | 25,729 |
| Water | 91 |
| Average B-factor | 64.4 |
| Protein | 64.5 |
| Water | 39.6 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.03 |
a Values in parentheses are for highest-resolution shell.
FIGURE 1.
Overall structure of human RNase H2. A and B, shown is the structure of the RNase H2 complex colored by subunits (yellow, A; magenta, B; blue, C). β-Strands are indicated with darker shades of the respective colors. C and D, shown is a close-up view of the interface between subunit A and the B/C dimer. The residues forming the interface are shown as sticks and labeled.
Aicardi-Goutières Syndrome Mutations in Human RNase H2
To date, 29 point mutations of RNase H2 have been reported in AGS patients. 8 were found in the A subunit, 14 in the B subunit, and 7 in the C subunit (12, 13, 25). Our structure permits all of these mutation sites to be located. One site, V185G, is located in a region that could be traced only in three of six complexes in the asymmetric unit and for which the electron density maps are poorly defined. Only 20 mutation sites were correctly placed in the mouse RNase H2 structure (Table 2).
TABLE 2.
Mutations found in AGS patients
Hs, human; Mm, mouse.
| Mutation | Mma | Hsb | Description | Proposed defect caused by the mutation |
|---|---|---|---|---|
| Subunit A | ||||
| D2Y+L3P | + | 3 | Surface (Asp), hydrophobic interaction (Leu) | Impaired target protein binding/structural stability |
| G37S | + | 6 | Located close to the active site, part of the GRG motif | Impaired substrate cleavage |
| R108W | + | 6 | Located at A/C interface | Impaired complex stability |
| R186W | + | 6 | Located close to the GRG motif | Impaired substrate cleavage |
| F230L | + | 6 | Located at the hydrophobic core | Impaired stability of the structure |
| R235Q | + | 6 | Stabilizes the conformation of the GRG motif | Impaired substrate cleavage |
| T240M | + | 6 | Interacts with the non-cleaved strand of the substrate | Impaired substrate cleavage |
| R291H | Mistraced fragment (mouse C:Trp-137 in this position) | 6 | Located at A/B/C interface | Impaired complex stability |
| Subunit B | ||||
| P43H | + | 6 | Located at the hydrophobic core/structural | Steric clash/impaired stability of the structure |
| L60R | + | 6 | Located at the hydrophobic core | Steric clash/impaired stability of the structure |
| W73L | + | 6 | Located at the hydrophobic core of B/C dimer | Impaired stability of the complex |
| G83S | + | 6 | Structural | Steric clash/impaired stability of the structure |
| H86R | + | 6 | Structural | Impaired stability of the structure |
| L138F | Mistraced fragment (mouse B:Ser-156 in this position) | 6 | Located at the hydrophobic core | Steric clash/impaired stability of the structure |
| S159I | Not visible | 6 | Surface | Impaired interaction with putative target proteins |
| K162T | + (Slightly displaced) | 6 | Surface | Impaired interaction with putative target proteins |
| T163I | + | 6 | Structural | Impaired stability of the structure |
| A177T | + | 6 | Structural/located close to the B/C interface | Steric clash/impaired stability of the complex |
| V183M | + | 6 | Located close to the A/B/C interface | Impaired complex stability |
| V185G | Not visible | 3 | Surface | |
| Y219H | Not visible | 6 | Located at the hydrophobic core close to A/B/C interface | Impaired stability of the structure/complex |
| S229P | Not visible | 6 | Surface | Impaired interaction with putative target proteins |
| Subunit C | ||||
| R13H | + | 6 | Surface | Impaired interaction with putative target proteins |
| D39Y | + | 6 | Located at the B/C interface | Impaired complex stability |
| R69W | + | 6 | Surface | |
| P76L | + | 6 | Structural | Impaired stability of the structure |
| P138L | Mistraced (mouse C:Asp-108 in this position) | 6 | Structural | Impaired stability of the structure |
| K143I | Mistraced fragment (mouse C:Pro-113 in this position) | 6 | Located at the A/B/C interface | Impaired complex stability |
| P151S | Mistraced fragment (mouse B:His-99 in this position) | 6 | Structural | Impaired stability of the structure |
a The + sign denotes residues correctly placed in the mouse model. Other residues are described in more detail.
b Number of Hs-RNase H2 complexes in the asymmetric unit in which the residue is observed.
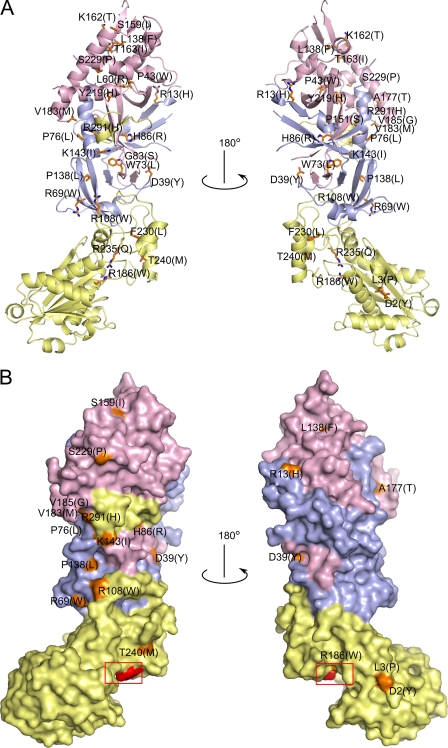
The mutant sites are located throughout the protein structure both on its surface and within the core (Fig. 3). Some of them are located in the vicinity of the subunit interface, and others are close to the active site and might be involved in substrate binding and hydrolysis. Based on the localization and environment of particular residues in the structure, the mutations can be classified into three groups as shown in Table 2. The proteins in each category may be compromised by different mechanisms, including (i) impairment of substrate binding/hydrolysis, (ii) impairment of structural integrity (residues in the hydrophobic core or at the subunit interfaces), and (iii) disruption of the interactions with potential interacting proteins (surface mutations).
FIGURE 3.
Aicardi-Goutières syndrome mutations in human RNase H2. A, shown is a schematic representation of the structure with the sites of mutations shown as orange sticks. B, shown is a surface representation of the structure showing the localization of surface-exposed mutation sites colored and labeled as in A. The red box indicates the position of the active site.
The first group is represented by an A:G37S mutation, which has been shown to impair the activity of human RNase H2 (12, 18, 26). As demonstrated by the substrate complex structure of T. maritima RNase H2 (Tm-RNase H2), Gly-37 is a part of the conserved GRG motif that is involved in specific interactions with the 2′-OH group of the (5′)RNA-DNA(3′) junction (17). We hypothesized that its mutation to serine would introduce steric clashes, leading to a change in conformation of the protein backbone that could in turn affect the interaction with the substrate (17). A neighboring mutation, A:R235Q, can have a similar effect. Arg-235 interacts with the GRG motif to stabilize it. Another mutation in the substrate interface region is T240M. As discussed below, Thr-240 is important for the binding of the non-cleaved strand, and its substitution to alanine greatly reduces the enzyme activity (Fig. 4C).
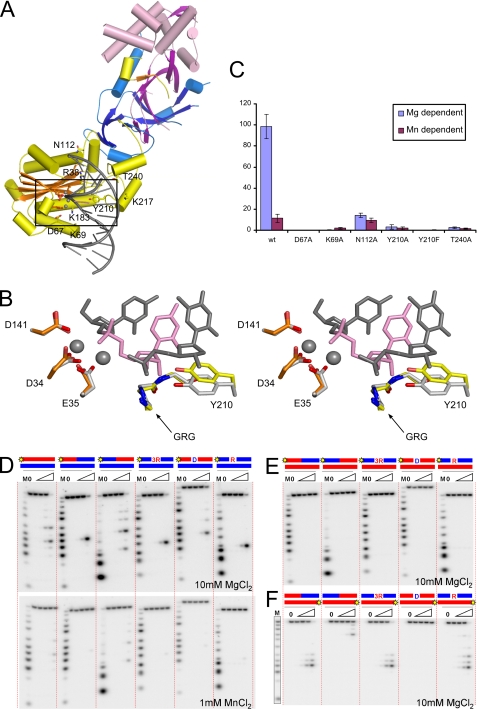
FIGURE 4.
Substrate recognition by human RNase H2. A, shown is a model of human RNase H2-substrate complex. The protein is colored as in Fig. 1. The active site residues (orange) and putative substrate binding residues (yellow) are shown as sticks and labeled. The positions of metal ions (gray spheres) and the substrate (gray with single ribonucleotide in pink) are inferred from the superposition of Tm-RNase H2-substrate complex (Protein Data Bank ID 3O3G) onto human RNase H2. B, shown is a stereo view of the human RNase H2 active site superimposed onto the active site of Tm-RNase H2. The active site residues of human RNase H2 are shown as orange sticks, and tyrosine 210 residue is shown as yellow sticks. Tm-RNase H2 is colored with shades of gray (active site, metal ions, and substrate), with the single ribonucleotide of the substrate shown in pink. C, activity of human RNase H2 variants with substitutions of postulated substrate binding residues is shown. The activity of the mutants in the presence of Mg2+ (blue) and Mn2+ (purple) ions was measured with poly(rA)/poly(dT) substrate. The results are shown as a percentage of the highest measured value for each experiment. Error bars represent the S.D. for each measurement. D, cleavage of duplexes formed by a DNA oligo hybridized to strands of various compositions by human RNase H2 is shown. The 5′-end 32P-labeled substrates (1 μm) indicated above the gel (RNA in red, DNA in blue, site of labeling shown as yellow star) were hydrolyzed with increasing concentrations of RNase H2 (11 pm, 110 pm, 1.1 nm, and 11 nm). The sequences and the major cleavage sites are summarized in supplemental Fig. 3. The lane without enzyme added is indicated with 0, and lanes marked with a triangle contained increasing amounts of protein. Twenty-microliter reaction mixtures were incubated at 37 °C for 15 min in the presence of 10 mm MgCl2 (upper panel) or 1 mm MnCl2 (lower panel). Products of the hydrolysis were analyzed on 20% Tris borate EDTA-urea polyacrylamide gels. The sizes of the products were measured based on molecular size markers indicated as M (products of digestion of 32P-labeled strands without complementary DNA by phosphodiesterase I). E and F, cleavage of duplexes formed by an RNA oligo hybridized to strands of various compositions in the presence of Mg2+.
The second type of mutations is exemplified by A:R108W. This arginine residue is located in a loop, which takes part in the formation of the interface between subunits A and C (Fig. 1D). It forms van der Waals contacts with C:Leu-134 and C:Glu-135 and a salt bridge with A:Glu-255. Mutation of Arg-108 to tryptophan likely disrupts those interactions and weakens the contact between subunits A and C. The mutation sites can also be buried within the core of the protein (e.g. B:W73L and B:Y219H). These mutations are likely to disrupt hydrophobic interactions and consequently destabilize the protein structure. They can also render the enzyme more susceptible to hydrolysis by proteases.
To test the effect of selected mutations on the stability of the human RNase H2, we prepared expression constructs for eight variants of the complex, all with substitutions in subunit B: P43H, L60R, W73L, G83S, H86R, S159I, T163I, and Y219H. Four of them could be produced in E. coli in soluble form and purified: W73L, G83S, H86R, and Y219H. The stability of these four variants was tested in a Thermofluor assay. The melting temperature of all four variants was lower by 5–7 °C compared with the wild type protein (Table 3), which indicates that the mutations perturbed the structure of the complex.
TABLE 3.
Thermal stability of human RNase H2 with selected AGS mutations (Thermofluor assay)
| Protein | Tm | ΔTm (compared with wild type) |
|---|---|---|
| °C | °C | |
| Wild type | 51.1 ± 0.1 | |
| B:W73L | 45.9 ± 0.3 | −5.2 |
| B:G83S | 45.7 ± 0.2 | −5.4 |
| B:H86R | 44.2 ± 0.1 | −6.9 |
| B:Y219H | 44.8 ± 0.1 | −6.3 |
The last group of mutations encompasses substitutions occurring on the protein surface. They often severely alter the chemical properties of the side chain (e.g. B:K162T), which significantly affects the local chemical properties of the protein surface. This could influence the interactions with as yet unidentified target proteins. In subunit B, three mutations cluster together (S159I, K162T, and T163I), suggesting that the region around them is important for the binding of putative protein partners.
Substrate Recognition by Human RNase H2
To better understand substrate recognition by human RNase H2, we built a model of the human protein in complex with nucleic acid (Fig. 4A) using the substrate complex structure of Tm-RNase H2 (17). Structures of the bacterial protein and the A subunit of the human protein are quite similar. When the Tm-RNase H2 substrate complex was superimposed on the structure of the human enzyme using the positions of 107 pairs of C-α atoms, the resulting root mean square deviation was 1.7 Å. In such a superimposition, the nucleic acid from the bacterial complex showed good complementarity to the surface of the human A subunit, except in the region of two loops (residues 141–145 and 209–214), which may change conformation upon substrate binding. Additionally, the active sites and the GRG 2′-OH-sensing motif of human and T. maritima RNases H2 superimpose very well, suggesting that the positions of the two catalytic metal ions and the mechanism of cleavage are conserved (Fig. 4B). The model shows that the B and C subunits are located away from the substrate interface and, therefore, cannot interact with the substrate in the immediate vicinity of the active site (Fig. 4A). This is consistent with the fact that AGS mutations found in the B and C subunits do not affect nuclease activity (9, 26).
We used our model to identify residues of the human protein that could be involved in substrate binding, and we verified their importance through site-directed mutagenesis. In Tm-RNase H2, Pro-187 forms a van der Waals interaction with the backbone of the non-cleaved strand. We previously hypothesized that its counterpart in the human enzyme, Thr-240, could play a similar role (17). Indeed, when we substituted Thr-240 with alanine, the activity of the human enzyme was greatly reduced on long hybrids (Fig. 4C) and also on short 12-mer substrates in the presence of Mg2+ ions (RNA/DNA, RNA-DNA/DNA, and DNA-RNA1-DNA/DNA) with the strongest inhibition on the regular hybrid and single-ribonucleotide oligo (not shown). Interestingly, Thr-240 is mutated to methionine in some AGS patients (13), which likely introduces large steric clashes with the nucleic acid, impairing its binding. Another residue involved in the binding of the non-cleaved strand in Tm-RNase H2 is Asn-81. When its human equivalent (Asn-112) was substituted with alanine, it led to a large decrease in magnesium-dependent activity, but manganese-dependent activity was largely unaffected (Fig. 4C). Based on the Tm-RNase H2 structure, we proposed that in the presence of Mn2+ ions, RNase H2 can act on substrates that are less well positioned at the active site (17). This suggests that the substitution of Asn-112 does not inhibit substrate binding but affects its positioning for cleavage. In our mutagenesis experiments, we also targeted the residues from a DSK motif (residues 67–69 in human RNase H2A) located in a loop in the vicinity of the active site. Although the DSK motif has a highly conserved sequence among RNases H2, it is poorly defined in all of the six copies of the complex in the human RNase H2 structure, which indicates that in the absence of substrate it is not well structured. In the Tm-RNase H2 complex, the motif interacts with the scissile phosphate and may participate in the active site formation (17). For the human enzyme, substitutions of Asp-67 and Lys-69 by alanine greatly reduced or abolished enzymatic activity on long hybrids (Fig. 4C) and short 12-mer substrates (not shown). Similar results were previously reported for Archeoglobus fulgidus RNase H2 (27), which confirms the general role played by the DSK motif in RNase H2 activity.
One important residue involved in the binding of the substrate in the vicinity of the active site (17) is the conserved tyrosine residue (Tyr-210 in human enzyme). Similar to bacterial and archaeal proteins (16, 17), Y210F and Y210A variants of the human enzyme lost most or all activity on long hybrids in the presence of both Mg2+ and Mn2+ ions (Fig. 4C). The two variants of the protein were also inactive on short 12-mer substrates (not shown). For Tm-RNase H2, the tyrosine side chain has been proposed to be an essential element selecting for RNA-DNA junctions and precluding the cleavage of RNA/DNA hybrids (17). However, eukaryotic RNases H2 can cleave RNA/DNA hybrids as well as RNA-DNA transitions (9). Therefore, the difference between eukaryotic and bacterial enzymes in their abilities to cleave RNA/DNA most likely stems from different positioning of the tyrosine residue. This tyrosine is absolutely conserved, yet it is quite striking that the region around it has a very different sequence in bacterial RNases H2 compared with archaeal/eukaryotic proteins (supplemental Fig. 2A). The region preceding the tyrosine residue also has a different structure in bacterial and archaeal/eukaryotic proteins. In the Tm-RNase H2 structure, it forms a short one-turn helix sandwiched between two other helices, whereas in human and archaeal structures, it forms an essentially straight segment (supplemental Fig. 2B). When human, archaeal, and bacterial RNases H2 are superimposed using the positions of the central β-sheet of the RNase H fold, the active sites overlap quite well, but the tyrosine residue in archaeal and human proteins is shifted by ∼2–4 Å away from the 2′-OH group of the RNA-DNA junction (supplemental Fig. 2C). This shift may lead to less discrimination against 2′-OH groups one nucleotide from the cleavage site, which is the likely reason why eukaryotic and archaeal enzymes can hydrolyze RNA/DNA hybrids that contain 2′-OH groups throughout the cleaved strand.
We tested the substrate preference of human RNase H2 on a number of RNA/DNA hybrids (Fig. 4, D–F). In the presence of Mg2+ ions, human RNase H2 cleaves the (5′)RNA-DNA(3′) junction, RNA/DNA, DNA-RNA/DNA, and RNA-DNA1-RNA/DNA substrates equally well. Supplemental Fig. 3A shows the preferred sites of cleavage for the different substrates. The activity in the presence of Mn2+ ions was generally much weaker, and the enzyme preferred RNA/DNA hybrids and DNA-RNA/DNA substrates over oligos with (5′)RNA-DNA(3′) junctions (Fig. 4D). Only at higher protein concentrations does human RNase H2 hydrolyze junction substrates in the presence of Mn2+ ions (data not shown). These results are consistent with previous studies of human RNase H2 (9, 12, 28). Similar substrate preferences were also reported for archaeal A. fulgidus (27) and Thermococcus kodakaraensis (28) RNases H2, but bacterial RNases H2 exhibit a different specificity. They prefer to cleave at the (5′)RNA-DNA(3′) junction, and they hydrolyze RNA/DNA hybrids efficiently only in the presence of Mn2+ with no cleavage observed with Mg2+ ions (2, 29).
Bacterial RNases H2 from E. coli and Thermus thermophilus can cleave substrates in which the non-cleaved strand contains RNA instead of DNA, with both enzymes cleaving the RNA-DNA/RNA duplex at the RNA-DNA junction (29). We tested the human enzyme for cleavage of duplexes formed by an RNA oligo hybridized to strands of various compositions (Fig. 4, E and F). The strand opposite to the RNA was not cleaved by human RNase H2 regardless of its composition, indicating that the human enzyme requires the DNA as the non-cleaved strand. The RNA strand itself was not cleaved when only a single deoxynucleotide was included in the complementary strand, but hydrolysis occurred when a DNA stretch was present (Fig. 4F and supplemental Fig. 3B).
Conclusions
We presented the structure of the human RNase H2 complex. We described a different tracing of subunits B and C of the complex compared with the previously reported mouse complex structure. This allowed us to map the positions of all of the reported AGS mutations on the structure. Based on their location, we classified them into three groups; (i) mutations affecting substrate binding/hydrolysis, (ii) mutations that prevent complex formation, and (iii) mutations that impair the interaction with other proteins. Based on our previous results, we also prepared a model of substrate recognition by human RNase H2 that allowed us to explain its substrate preference. Significant evidence has accumulated recently indicating the crucial role of RNase H2 in the removal of single ribonucleotides from the DNA and maintaining the stability of the genome. Our results are an important step toward full elucidation of the biochemical properties of this interesting enzyme.
Acknowledgments
We thank Monika Rychlik and Jadwiga Dyttus for excellent technical assistance, the staff of beamline 23-2 at European Synchrotron Radiation Facility for assistance with data collection, and Dr. Karolina Górecka for help with data collection.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of the Eunice Kennedy Shriver NICHD. This work was also supported by Wellcome Trust International Senior Fellowship 081760. The access to the European Synchrotron Radiation Facility was financed by the Polish Ministry of Science and Higher Education (Project ESRF/73/2006).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1 and 2.
The atomic coordinates and structure factors (code 3PUF) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AGS
- Aicardi-Goutières syndrome
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tm
- T. maritima
- poly(rA)/poly(dT)
- polyadenylic acid/poly-deoxyribothymidylic acid.
REFERENCES
- 1. Tadokoro T., Kanaya S. (2009) FEBS J. 276, 1482–1493 [DOI] [PubMed] [Google Scholar]
- 2. Ohtani N., Haruki M., Morikawa M., Crouch R. J., Itaya M., Kanaya S. (1999) Biochemistry 38, 605–618 [DOI] [PubMed] [Google Scholar]
- 3. Nowotny M. (2009) EMBO Rep. 10, 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rydberg B., Game J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16654–16659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qiu J., Qian Y., Frank P., Wintersberger U., Shen B. (1999) Mol. Cell. Biol. 19, 8361–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arudchandran A., Cerritelli S., Narimatsu S., Itaya M., Shin D. Y., Shimada Y., Crouch R. J. (2000) Genes Cells 5, 789–802 [DOI] [PubMed] [Google Scholar]
- 8. Jeong H. S., Backlund P. S., Chen H. C., Karavanov A. A., Crouch R. J. (2004) Nucleic Acids Res. 32, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chon H., Vassilev A., DePamphilis M. L., Zhao Y., Zhang J., Burgers P. M., Crouch R. J., Cerritelli S. M. (2009) Nucleic Acids Res. 37, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frank P., Braunshofer-Reiter C., Wintersberger U., Grimm R., Büsen W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12872–12877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eder P. S., Walder J. A. (1991) J. Biol. Chem. 266, 6472–6479 [PubMed] [Google Scholar]
- 12. Crow Y. J., Leitch A., Hayward B. E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., Baumann C., Baxter P., Bertini E., Chandler K. E., Chitayat D., Cau D., Déry C., Fazzi E., Goizet C., King M. D., Klepper J., Lacombe D., Lanzi G., Lyall H., Martínez-Frías M. L., Mathieu M., McKeown C., Monier A., Oade Y., Quarrell O. W., Rittey C. D., Rogers R. C., Sanchis A., Stephenson J. B., Tacke U., Till M., Tolmie J. L., Tomlin P., Voit T., Weschke B., Woods C. G., Lebon P., Bonthron D. T., Ponting C. P., Jackson A. P. (2006) Nat. Genet. 38, 910–916 [DOI] [PubMed] [Google Scholar]
- 13. Rice G., Patrick T., Parmar R., Taylor C. F., Aeby A., Aicardi J., Artuch R., Montalto S. A., Bacino C. A., Barroso B., Baxter P., Benko W. S., Bergmann C., Bertini E., Biancheri R., Blair E. M., Blau N., Bonthron D. T., Briggs T., Brueton L. A., Brunner H. G., Burke C. J., Carr I. M., Carvalho D. R., Chandler K. E., Christen H. J., Corry P. C., Cowan F. M., Cox H., D'Arrigo S., Dean J., De Laet C., De Praeter C., Dery C., Ferrie C. D., Flintoff K., Frints S. G., Garcia-Cazorla A., Gener B., Goizet C., Goutieres F., Green A. J., Guet A., Hamel B. C., Hayward B. E., Heiberg A., Hennekam R. C., Husson M., Jackson A. P., Jayatunga R., Jiang Y. H., Kant S. G., Kao A., King M. D., Kingston H. M., Klepper J., van der Knaap M. S., Kornberg A. J., Kotzot D., Kratzer W., Lacombe D., Lagae L., Landrieu P. G., Lanzi G., Leitch A., Lim M. J., Livingston J. H., Lourenco C. M., Lyall E. G., Lynch S. A., Lyons M. J., Marom D., McClure J. P., McWilliam R., Melancon S. B., Mewasingh L. D., Moutard M. L., Nischal K. K., Ostergaard J. R., Prendiville J., Rasmussen M., Rogers R. C., Roland D., Rosser E. M., Rostasy K., Roubertie A., Sanchis A., Schiffmann R., Scholl-Burgi S., Seal S., Shalev S. A., Corcoles C. S., Sinha G. P., Soler D., Spiegel R., Stephenson J. B., Tacke U., Tan T. Y., Till M., Tolmie J. L., Tomlin P., Vagnarelli F., Valente E. M., Van Coster R. N., Van der Aa N., Vanderver A., Vles J. S., Voit T., Wassmer E., Weschke B., Whiteford M. L., Willemsen M. A., Zankl A., Zuberi S. M., Orcesi S., Fazzi E., Lebon P., Crow Y. J. (2007) Am. J. Hum. Genet. 81, 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muroya A., Tsuchiya D., Ishikawa M., Haruki M., Morikawa M., Kanaya S., Morikawa K. (2001) Protein Sci. 10, 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai L., Yokota H., Hung L. W., Kim R., Kim S. H. (2000) Structure Fold Des. 8, 897–904 [DOI] [PubMed] [Google Scholar]
- 16. Chapados B. R., Chai Q., Hosfield D. J., Qiu J., Shen B., Tainer J. A. (2001) J. Mol. Biol. 307, 541–556 [DOI] [PubMed] [Google Scholar]
- 17. Rychlik M. P., Chon H., Cerritelli S. M., Klimek P., Crouch R. J., Nowotny M. (2010) Mol. Cell 40, 658–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shaban N. M., Harvey S., Perrino F. W., Hollis T. (2010) J. Biol. Chem. 285, 3617–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W. (1997) Methods in Enzymol. (Carter C. W., Sweet R. M. eds) pp. 307–326, Academic Press, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 20. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) CCP4 Newsletter 42, contribution 8 [Google Scholar]
- 22. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23. Gaidamakov S. A., Gorshkova I. I., Schuck P., Steinbach P. J., Yamada H., Crouch R. J., Cerritelli S. M. (2005) Nucleic Acids Res. 33, 2166–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nettleship J. E., Brown J., Groves M. R., Geerlof A. (2008) Methods Mol. Biol. 426, 299–318 [DOI] [PubMed] [Google Scholar]
- 25. Ramantani G., Kohlhase J., Hertzberg C., Innes A. M., Engel K., Hunger S., Borozdin W., Mah J. K., Ungerath K., Walkenhorst H., Richardt H. H., Buckard J., Bevot A., Siegel C., von Stülpnagel C., Ikonomidou C., Thomas K., Proud V., Niemann F., Wieczorek D., Häusler M., Niggemann P., Baltaci V., Conrad K., Lebon P., Lee-Kirsch M. A. (2010) Arthritis Rheum. 62, 1469–1477 [DOI] [PubMed] [Google Scholar]
- 26. Perrino F. W., Harvey S., Shaban N. M., Hollis T. (2009) J. Mol. Med. 87, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chai Q., Qiu J., Chapados B. R., Shen B. (2001) Biochem. Biophys. Res. Commun. 286, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 28. Rohman M. S., Koga Y., Takano K., Chon H., Crouch R. J., Kanaya S. (2008) FEBS J. 275, 4836–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohtani N., Tomita M., Itaya M. (2008) FEBS J. 275, 5444–5455 [DOI] [PubMed] [Google Scholar]