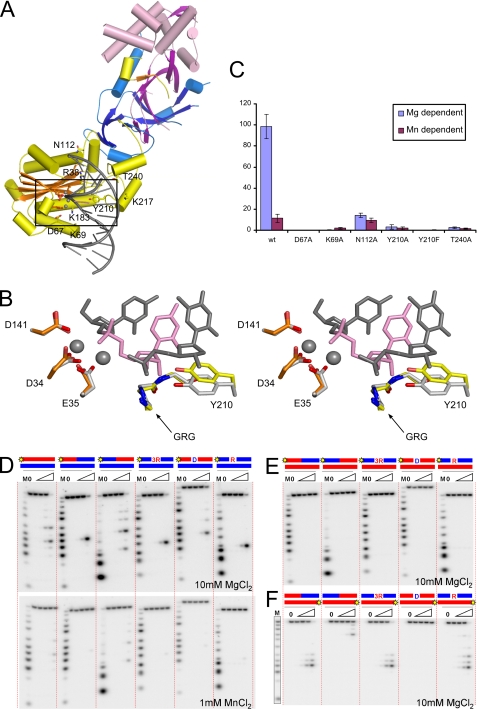
FIGURE 4.
Substrate recognition by human RNase H2. A, shown is a model of human RNase H2-substrate complex. The protein is colored as in Fig. 1. The active site residues (orange) and putative substrate binding residues (yellow) are shown as sticks and labeled. The positions of metal ions (gray spheres) and the substrate (gray with single ribonucleotide in pink) are inferred from the superposition of Tm-RNase H2-substrate complex (Protein Data Bank ID 3O3G) onto human RNase H2. B, shown is a stereo view of the human RNase H2 active site superimposed onto the active site of Tm-RNase H2. The active site residues of human RNase H2 are shown as orange sticks, and tyrosine 210 residue is shown as yellow sticks. Tm-RNase H2 is colored with shades of gray (active site, metal ions, and substrate), with the single ribonucleotide of the substrate shown in pink. C, activity of human RNase H2 variants with substitutions of postulated substrate binding residues is shown. The activity of the mutants in the presence of Mg2+ (blue) and Mn2+ (purple) ions was measured with poly(rA)/poly(dT) substrate. The results are shown as a percentage of the highest measured value for each experiment. Error bars represent the S.D. for each measurement. D, cleavage of duplexes formed by a DNA oligo hybridized to strands of various compositions by human RNase H2 is shown. The 5′-end 32P-labeled substrates (1 μm) indicated above the gel (RNA in red, DNA in blue, site of labeling shown as yellow star) were hydrolyzed with increasing concentrations of RNase H2 (11 pm, 110 pm, 1.1 nm, and 11 nm). The sequences and the major cleavage sites are summarized in supplemental Fig. 3. The lane without enzyme added is indicated with 0, and lanes marked with a triangle contained increasing amounts of protein. Twenty-microliter reaction mixtures were incubated at 37 °C for 15 min in the presence of 10 mm MgCl2 (upper panel) or 1 mm MnCl2 (lower panel). Products of the hydrolysis were analyzed on 20% Tris borate EDTA-urea polyacrylamide gels. The sizes of the products were measured based on molecular size markers indicated as M (products of digestion of 32P-labeled strands without complementary DNA by phosphodiesterase I). E and F, cleavage of duplexes formed by an RNA oligo hybridized to strands of various compositions in the presence of Mg2+.