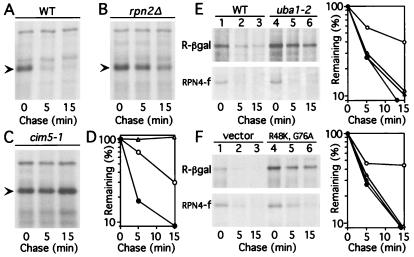
Figure 2.
RPN4 is a short-lived protein degraded by a proteasome-dependent pathway. (A–C) Pulse–chase analysis of C-terminally tagged RPN4 (RPN4-flag) that was expressed from the induced PCUP1 promoter and low-copy plasmid either in wild-type (RPN4) strain JD52 (A), in a congenic rpn2Δ mutant (B), or in a cim5–1 mutant (C). Cells were labeled at 28°C and chased at 28°C in A and B, and at 37°C in C. The kinetics of RPN4 degradation in JD52 were similar at 28 and 37°C (data not shown). Arrowheads indicate the band of RPN4-flag. (D) Quantitation of the patterns in A–C, by using PhosphorImager (Molecular Dynamics) ●, wild-type cells. ○, rpn2Δ cells. ▵, cim5–1 cells. (E) Pulse–chase analyses of Arg-βgal (R-βgal), derived from Ub-Arg-βgal (3, 14), and of RPN4-flag (expressed as in A) in uba1–2 (37) and wild-type cells. Quantitation: ● and ○, Arg-βgal in wild-type and uba1–2 cells, respectively. ▴ and ▵, RPN4-flag in the same strains. (F) The same test proteins in wild-type cells and cells overexpressing UbK48R, G76A, with quantitation on the right; same designations. The rpn2Δ locus, in the strain AVY302 (see Materials and Methods), was a disruption allele. It was produced through the integration of URA3 at the HindIII site of RPN2 (codon 146) and was identical to the rpn2∷URA3 allele described by Yokota et al. (36). The phenotypes of AVY302 and the previously described strain (36) were similar as well (data not shown). Two other studies reported that rpn2Δ cells were inviable (4, 56). The disruption of RPN2 in these works was carried out with either TRP1 or ADE2, by using the RPN2 BglII site at codon 38. It remains to be determined whether different integration sites account for different phenotypes described for rpn2Δ strains.