Table 3. ML estimates and 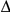 AIC values of the present models for the 1-PAM amino acid substitution matrices of JTT, WAG, and LG, and the 1-PAM codon substitution matrix of KHG.
AIC values of the present models for the 1-PAM amino acid substitution matrices of JTT, WAG, and LG, and the 1-PAM codon substitution matrix of KHG.
| JTT | WAG | LG | KHG | |||||||
| (codon) | ||||||||||
| id | parameter | ML–87a | ML–91a | ML–94 | ML–87a | ML–91a | ML–94 | ML–91a | ML–94 | ML–200 |
| no. | ||||||||||
| 0 |
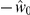
|
N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 1 |
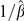
|
N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 2 |
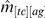
|
(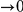 ) ) |
0.637 | 0.662 | (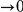 ) ) |
1.28 | 1.29 | 1.08 | 1.19 | 0.939 |
| 3 |
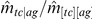
|
0.0919 | 1.57 | 1.59 | 0.746 | 1.70 | 1.69 | 1.85 | 1.81 | 0.843 |
| 4 |
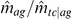
|
1.77 | 1.14 | 1.15 | 1.98 | 1.32 | 1.31 | 1.23 | 1.21 | 0.945 |
| 5 |
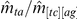
|
0.0293 | 0.729 | 0.730 | 0.0477 | 0.791 | 0.784 | 0.676 | 0.682 | 1.52 |
| 6 |
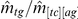
|
3.21 | 0.940 | 0.950 | 3.64 | 1.04 | 1.01 | 1.07 | 1.07 | 0.554 |
| 7 |
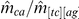
|
0.719 | 1.19 | 1.18 | 0.110 | 1.23 | 1.23 | 1.28 | 1.25 | 0.573 |
| 8 |
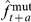
|
0.408 | 0.459 | 0.446 | 0.372 | 0.367 | 0.392 | 0.388 | 0.403 | 0.497 |
| 9 |
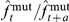
|
0.113 | 0.501 | 0.522 | 0.234 | 0.587 | 0.513 | 0.450 | 0.439 | 0.513 |
| 10 |
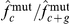
|
0.698 | 0.429 | 0.436 | 0.425 | 0.479 | 0.471 | 0.427 | 0.383 | 0.470 |
| 11 |
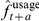
|
0.0682 | (0.5) | 0.483 | 0.0669 | (0.5) | 0.221 | (0.5) | 0.447 | NA |
| 12 |
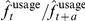
|
0.461 | (0.5) | 0.491 | 0.330 | (0.5) | 0.429 | (0.5) | 0.555 | NA |
| 13 |
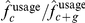
|
0.386 | (0.5) | 0.558 | 0.310 | (0.5) | 0.306 | (0.5) | 0.249 | NA |
| 14 |
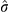
|
27.3 | 0.738 | 0.740 | 43.3 | 0.905 | 0.840 | 0.415 | 0.395 |
|
|
0.334 | 0.0243 | 0.0246 | 0.317 | 0.0223 | 0.0207 | 0.0246 | 0.0240 | 0.0240 | |
| #parameters | 107 | 111 | 114 | 107 | 111 | 114 | 111 | 114 | 261 | |
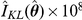 b
b
|
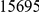
|
638 |
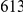
|
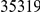
|
1903 |
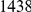
|
2771 | 2335 | 269946 | |
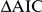 c
c
|
2072.0 | 297.5 | 300.6 | 1370.8 | 284.3 | 275.1 | 782.5 | 700.4 | unknown | |
| Ratio of substitution rates | ||||||||||
| per codon | ||||||||||
| the total base/codon | 1.28 | 1.35 | 1.35 | 1.38 | 1.53 | 1.52 | 1.38 | 1.39 | 1.29 | |
| (1.29)d | ||||||||||
| transition/transversion | 0.464 | 1.08 | 1.08 | 0.482 | 0.932 | 0.806 | 1.18 | 1.20 | 0.764 | |
| (0.765)d | ||||||||||
| nonsynonymous/synonymouse | 1.13 | 1.37 | 1.34 | 1.57 | 2.07 | 2.40 | 1.05 | 1.20 | 0.726 | |
| (0.723)d | ||||||||||
| Ratio of substitution rates | ||||||||||
per codon for 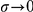
|
||||||||||
| total base/codon | 1.0 | 1.22 | 1.22 | 1.0 | 1.38 | 1.40 | 1.31 | 1.33 | 1.29 | |
| transition/transversion | 0.101 | 1.21 | 1.22 | 0.647 | 1.11 | 0.932 | 1.31 | 1.35 | 0.764 | |
| nonsynonymous/synonymouse | 0.0644 | 1.04 | 1.02 | 0.138 | 1.50 | 1.79 | 0.853 | 0.889 | 0.726 | |
| Ratio of substitution rates per | ||||||||||
codon for 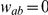 and and 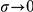
|
||||||||||
| total base/codon | 1.0 | 1.45 | 1.46 | 1.0 | 1.72 | 1.74 | 1.67 | 1.71 | 1.51 | |
| transition/transversion | 0.0605 | 0.829 | 0.831 | 0.499 | 0.933 | 0.849 | 0.992 | 0.981 | 0.427 | |
| nonsynonymous/synonymouse | 11.3 | 5.58 | 5.74 | 11.1 | 8.68 | 11.1 | 7.45 | 8.46 | 6.81 | |
aIf the value of a parameter is parenthesized, the parameter is not variable but fixed to the value specified.
b
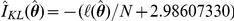 for JTT,
for JTT, 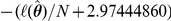 for WAG,
for WAG, 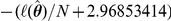 for LG, and
for LG, and 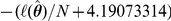 for KHG; see text for details.
for KHG; see text for details.
c
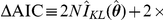 #parameters with
#parameters with 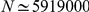 for JTT,
for JTT, 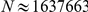 for WAG,
for WAG, 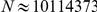 for LG, and the value of
for LG, and the value of 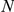 is unknown for KHG; see text for details.
is unknown for KHG; see text for details.
dThe value in the parenthesis corresponds to the one for the KHG codon substitution probability matrix.
eNote that these ratios are not the ratios of the rates per site but per codon; see text for details.
