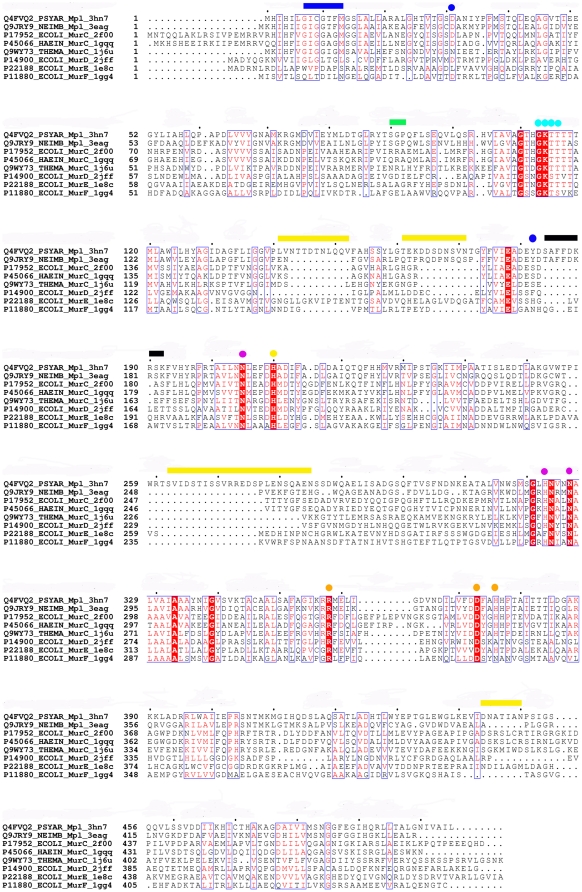
Figure 2. Sequence comparison of Mpl to MurC-F enzymes.
Multiple sequence alignment (CLUSTALW [55]) of Mpl with MurC-F, rendered using ESPRIPT [56]. The pairwise sequence identity between Mpl and MurC is ∼21–25%, and is less between Mpl and MurD-F, ∼15–17%. The PaMpl segment of residues 184–192 (black bar), comprised of the sequence motif (S/T)AFFDKRSK, is not present in Mur ligases and may be functionally important in Mpl. This motif packs against residues 90–92 (green bar), which are unique to and conserved in Mpl proteins (from analysis of 17 annotated Mpl enzymes in the UniProt database, August 2009, data not shown). Residues 7–13 constituting the GI(C/G)GTFM motif (blue bar) and Asp31 (blue circle) are likely to interact with UDP. Tyr182 (blue circle) is unique to Mpl and may be involved in substrate recognition. Residues that are likely to be important in ATP binding are: Asn205, His323 and Asn327 (adenine-binding, magenta circles); Asp373 (ribose-binding, orange circle); and Gly113, Lys114, Thr115, Thr116, Arg358 and His376 (tri-phosphate binding, cyan and orange circles). Metal-binding residues are likely to be Thr115, Glu178 and His210. The Psychrobacter Mpl enzymes have several insertions (residues 141–152, 161–171, 262–285 and 444–450, yellow bars), which may be related to cell wall recycling in these bacteria that survive in permafrost conditions.