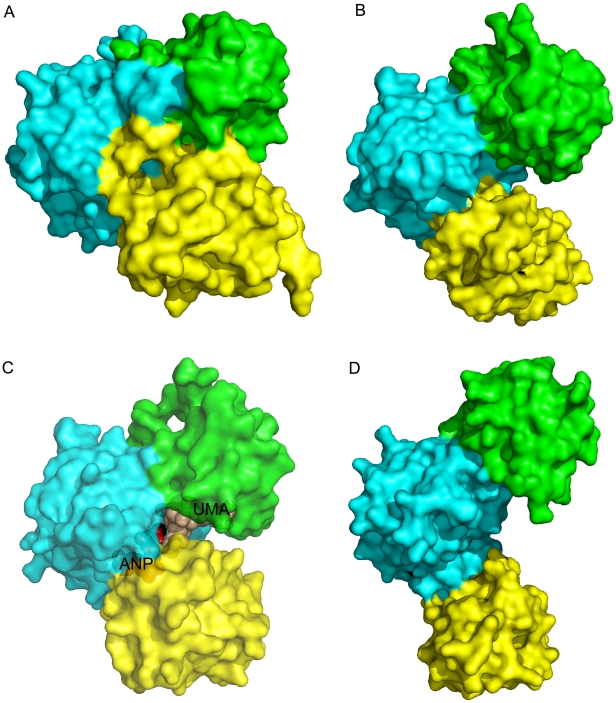
Figure 6. Surface representation of different domain dispositions in Mur family structures.
Crystal structures of: (A) apo-PaMpl, (B) EcMurC with bound Mg2+, (C) HiMurC bound to substrates UDP-MurNAc-L-Ala (UMA) and AMPPNP (ANP), and metal; and (D) apo-EcMurF (on slightly smaller scale compared to A–C as more extended conformation). For all proteins, ND is in green, MD is in cyan and CD is in yellow. These examples illustrate the conformational variability of these Mur enzymes. All molecules are in the same orientation based on their superimposition on the PaMpl MD. The CD of the apo-PaMpl is rotated 30° with respect to ND and MD compared to the MurC structure. The PaMpl domains may open up during substrate binding and close during catalysis.