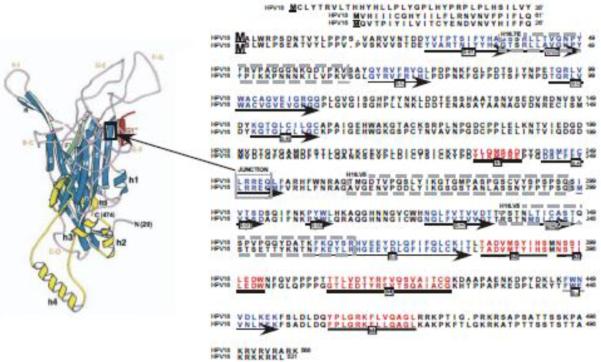
Figure 2.
Position of the Chimeric Junction, HPV16 L1-Reactive Conformation-Dependent MAb Epitopes, Structural Motifs and Sequence Homology of HPV18 and HPV16 L1 Proteins. The L1 protein amino acids sequences for HPV18 and HPV16 are aligned. Sequence numbering begins with the `consensus' methionines used for producing VLPs, PsV, and QV (Chen et al., 2010). The extended N-terminal sequences present in native viruses are given prime numbers, 1'–61' for HPV18 and 1'–26' for HPV16. Structural motifs are marked underneath the appropriate sequences (Chen et al., 2000) by arrows for ß-sheets and bolden lines for helices. HPV16 L1-reactive conformation-dependent MAb epitopes (Christensen et al., 2001; Culp et al., 2007) are outlined by dashed line boxes. The chimeric junction, LRREQ amino acids 250–254 is outlined by a solid box.