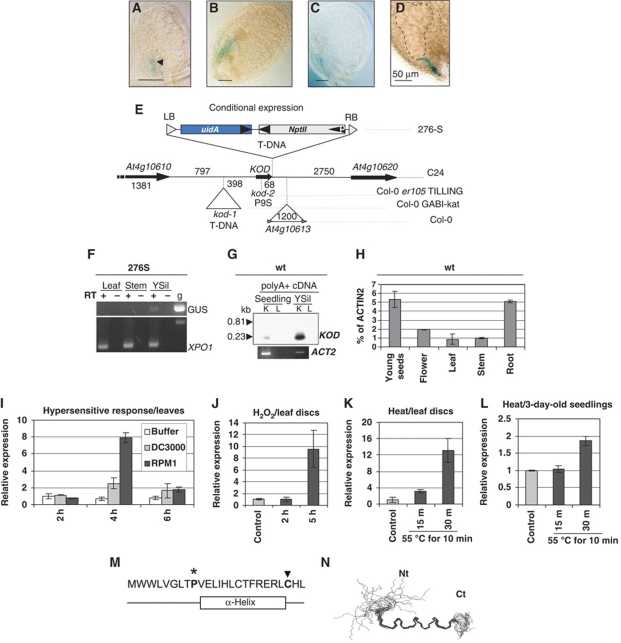
Figure 1.
Identification and expression profile of KOD. (A–E) Trap line and KOD locus. GUS expression pattern in line 276S. Arrowhead shows the boundary between the suspensor and the embryo proper. Stages of embryo development (A) globular, (B) late globular, (C) heart, (D) torpedo. Bars=50 μm. (E) Locus and structure of the promoter trap T-DNA in 276S (ecotype C24). At4g10613: LINE retrotransposon in Col-0; kod-1: T-DNA position in the GABI-Kat line. P9S, non-synonymous substitution in kod-2 (TILLING). uidA, β-glucuronidase gene. DNA sizes are given in bp. (F–L) Expression analysis of KOD. (F) RT–PCR analysis on tissues from line 276S using primer oexTi15 in KOD and oGUSj in the GUS gene. ‘GUS’ indicates the transcriptional fusion between KOD and the GUS sequence. XPO1, a ubiquitous gene (At5g17020) as a control. g, genomic DNA; YSil, young silique. (G) RT–PCR on polyA+ cDNA of WT, blotted and probed with a radiolabelled KOD ORF. K, Primer pair (oexTi15; oSUPR1) amplifying the KOD transcript (0.23 kb); L, primer pair (oTi05; oSUPR1) with oTi05 outside the transcript (0.81 kb). Expected sizes are shown; ACT2, Actin2 control. (H) KOD expression in Col-0 wild-type tissues using Taqman QRT–PCR. Actin2 is used as the reference gene. (I) KOD expression using Taqman QRT–PCR in WT leaves following infiltration with 10 mM MgCl2 (Buffer), P. syringae pv. tomato DC3000 wild-type (virulent) or expressing AvrRPM1 (avirulent). 18S is used as the reference gene. Each sample was measured in triplicate and is shown relative to expression at 2 h post-inoculation (hpi). (J) KOD expression using Taqman QRT–PCR in WT leaf discs floated on SDW (control) or 30 mM H2O2 for 2 and 5 h. 18S is used as the reference gene. Each sample was measured in triplicate and is shown relative to expression in the control. (K) KOD expression using Taqman QRT–PCR in WT leaf discs floated on SDW at RT (control) or 15 and 30 min after heat shock at 55°C for 10 min. 18S is used as the reference gene. Each sample was measured in triplicate and is shown relative to expression of control. (L) KOD expression using Taqman QRT–PCR in WT 3-day-old seedlings floated on SDW at RT (control) or 15 and 30 min after heat shock at 55°C for 10 min. 18S is used as the reference gene. Each sample was measured in triplicate and is shown relative to expression of control. (M) Amino-acid sequence of KOD and position of α-helix, see Supplementary Figure S1. Star and arrow highlight 2 aa required for full functionality of KOD, see Figure 5C. (N) Overlay of 22 calculated structures from NMR spectra. Nt, N-terminus; Ct, C-terminus. Residues 9–21-fold into an α-helix, whereas the N-terminal region is disordered.