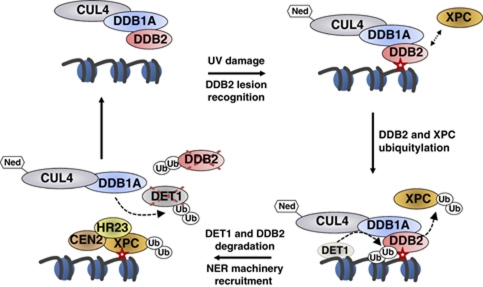
Figure 7.
A working model for DET1 cooperation with CUL4–DDB1DDB2 in DNA repair. UV-DNA photoproduct (red star) is detected and bound by DDB2, which focalizes neddylated (Ned) CUL4–DDB1 ubiquitin ligase on the damaged region. In mammals, XPC is recruited and poly-ubiquitylated by CUL4–DDB1DDB2, thereby enhancing its binding to DNA. DDB2 protein is subsequently ubiquitylated and degraded by a CUL4–DDB1- and DET1-dependent mechanism. This involves the formation of a transient large DET1 complex presumably containing CUL4 in addition to DDB1, which is a stable partner of DET1 through direct protein–protein interaction. DET1 would then also be targeted by a CUL4–DDB1 ubiquitin ligase for degradation. Proteolysis of DDB2 and DET1 may allow their eviction from chromatin to facilitate recruiting the NER machinery, which is initiated by binding of the heterotrimeric XPC–HR23–CEN2 (plant RAD4-RAD23-CEN2) factor onto the lesion. DET1 has the capacity to bind histone H2B, represented here on an adjacent nucleosome for simplicity, but whether it acts as a soluble or histone-bound factor for DDB2 degradation remains to be determined.