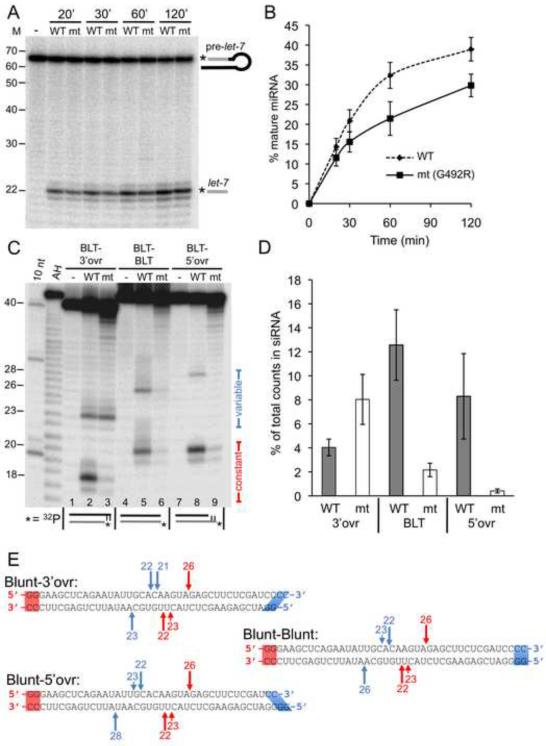
Figure 2.
The helicase domain of C. elegans Dicer is required for cleavage of dsRNA with blunt or 5' overhanging termini, but not 3' overhanging termini. (A) PhosphorImage showing production of mature let-7 miRNA (let-7), from 5' 32P-end-labeled (*) pre-let-7, incubated for indicated times without (−), or with, WT or G492R mutant (mt) C. elegans extract in extract cleavage buffer (12 mM MgOAc; Experimental Procedures). M, positions of RNA decade ladder and 22 nt miRNA. (B) Data from multiple assays as in (A) were quantified to determine % miRNA (100 × [radioactivity in miRNA band/total radioactivity in lane]); miRNA band included radioactivity in major and two immediately adjacent bands, but plots did not differ significantly when only predominant band was monitored. Data points show average values; error bars, standard error of the mean; n=2. (C) PhosphorImage of reaction products generated after 60 min incubation of 40/42 dsRNAs with indicated termini, without (−), or with, WT or G492R mutant (mt) extract in extract cleavage buffer (10 mM MgOAc; Experimental Procedures). As cartooned, 40/42 dsRNAs were 5' 32P-end-labeled on bottom strand. Products generated by Dicer measuring from constant or variable termini are indicated. Marked lengths (left) were determined with reference to AH ladders,10 nt RNA ladders (10 nt), and T1 ribonuclease ladders (Figure S1). (D) Data from multiple assays as in (C) were quantified to compare cleavage from three types of termini. % siRNA = 100 × (radioactivity in siRNA band/total radioactivity in lane); radioactivity in siRNA band included predominant and immediately adjacent bands if present. % siRNA values for BLT end processing included constant ends of 3'-ovr and 5'-ovr dsRNAs as well as both termini of BLT-BLT dsRNAs. Error bars, standard error of the mean (n≥6); mt is G492R. (E) Schematic of observed cleavage sites in 40/42 dsRNAs reacted as in (C) with extracts of wildtype C. elegans. Sites were determined using data for 40/42 dsRNAs labeled on the top (Figure S1) or bottom (Figure 2C) strand. Length in nts is shown above arrows marking major (long arrows) and minor (short arrows) cleavage with respect to Dicer measurement from constant blunt end (red) or variable end (blue).