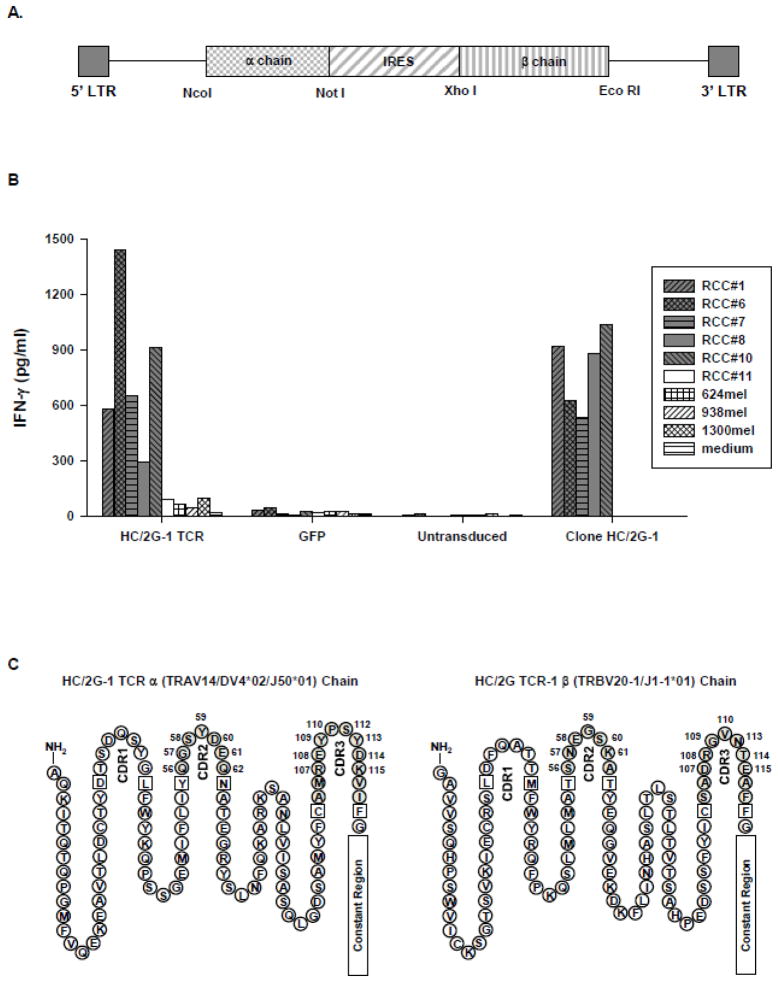
Figure 1.
Evaluation of the retrovirus encoding the native HC/2G-1 TCR. A. Schematic of retroviral vector containing HC/2G-1 TCR α and β chains separated by internal ribosomal entry site (IRES). NcoI, NotI, XhoI and EcoRI are the restriction endonuclease sites. B. IFN-γ production by PBL transduced with the HC/2G-1 TCR. Allogeneic PBL were stimulated with anti-CD3 (50ng/ml) for 2 days and transduced twice with retrovirus encoding HC/2G-1 TCR at 0.5 × 106 cells/well in a 24-well plate. PBL retrovirally transduced with green fluorescence protein (GFP) was used as the control. 3 days after transduction, transduced cells (1 × 105) were co-cultured with 5 x104 renal tumor cells (RCC #1, 6, 7. 8, 10 and 11) or melanoma cells (624mel, 938mel and 1300mel). After overnight incubation, the supernatant was harvested and IFN-γ production was measured. GFP-transduced and untransduced PBL were used as controls. The clone HC/2G-1 (1 x104/well) was also included in the experiment. C. Primary structure of the HC/2G-1 TCR α and β chains with CDRs (complementarity determining region) noted. Numbered AA positions are noted to identify subsequent amino acid (AA) substitutions (Note: not all consensus CDR aa positions are present after TCR recombination).