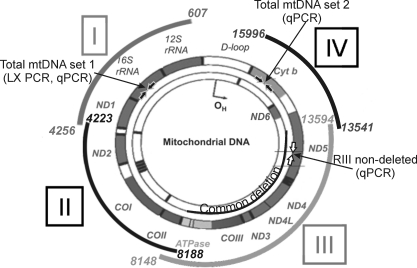
Figure 2.
Primer locations within the mt genome. Primers were designed to amplify discrete regions (I-IV) of the mt genome. The area spanned by each region is shown as a curved segment outside the mt genome, and base pair locations are provided. The primer set amplifying a 191-bp region that gives a measure of the total amount of mtDNA (total mtDNA set 1) is shown within the 16S rRNA gene (filled block arrows). A second set of primers amplifying a 222-bp region in the Cyt b gene provided a second measure of total mtDNA content for this study (total mtDNA primer set 2, solid block arrows). The RIII-ND primer set, which binds within the CD, is shown in the ND5 gene (open block arrows). The 4977-bp CD is shown within the mt genome. 12S and 16S rRNA are genes for mt ribosomal RNA. Genes for electron transport proteins are ND1–6, COI-III, ATPase 6, ATPase 8, and Cyt b. D-loop is the noncoding region. The schematic of the mt genome was modified with permission from Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Gen. 2005;6:389–402.