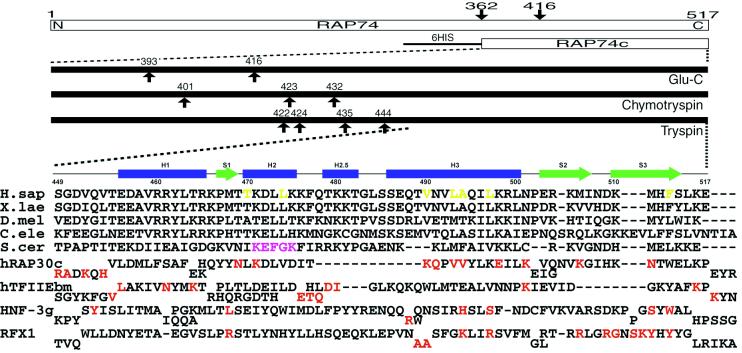
Figure 1.
(Upper) Summary of domain mapping within human RAP74 by using limited proteolysis combined with mass spectrometry. Arrows indicate cleavage sites observed within minutes to hours. (Lower) Sequence alignments of the C-terminal regions of RAP74 from human (H.sap), Xenopus laevis, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae. Secondary structure elements observed in the hRAP74cc crystal structure are also shown. Structure-based sequence alignments of the C-terminal domain of human RAP30, the middle domain of TFIIEβ, and the DNA-binding domains of HNF-3γ and RFX1 are given below. Red letters denote DNA-binding residues identified by x-ray crystallography or NMR. Magenta and yellow letters denote putative FCP1-binding regions identified in yeast and residues constituting the hydrophobic concave surface of hRAP74cc, respectively.