Abstract
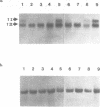
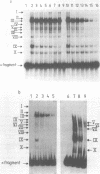
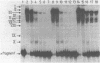
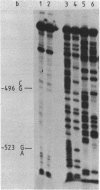
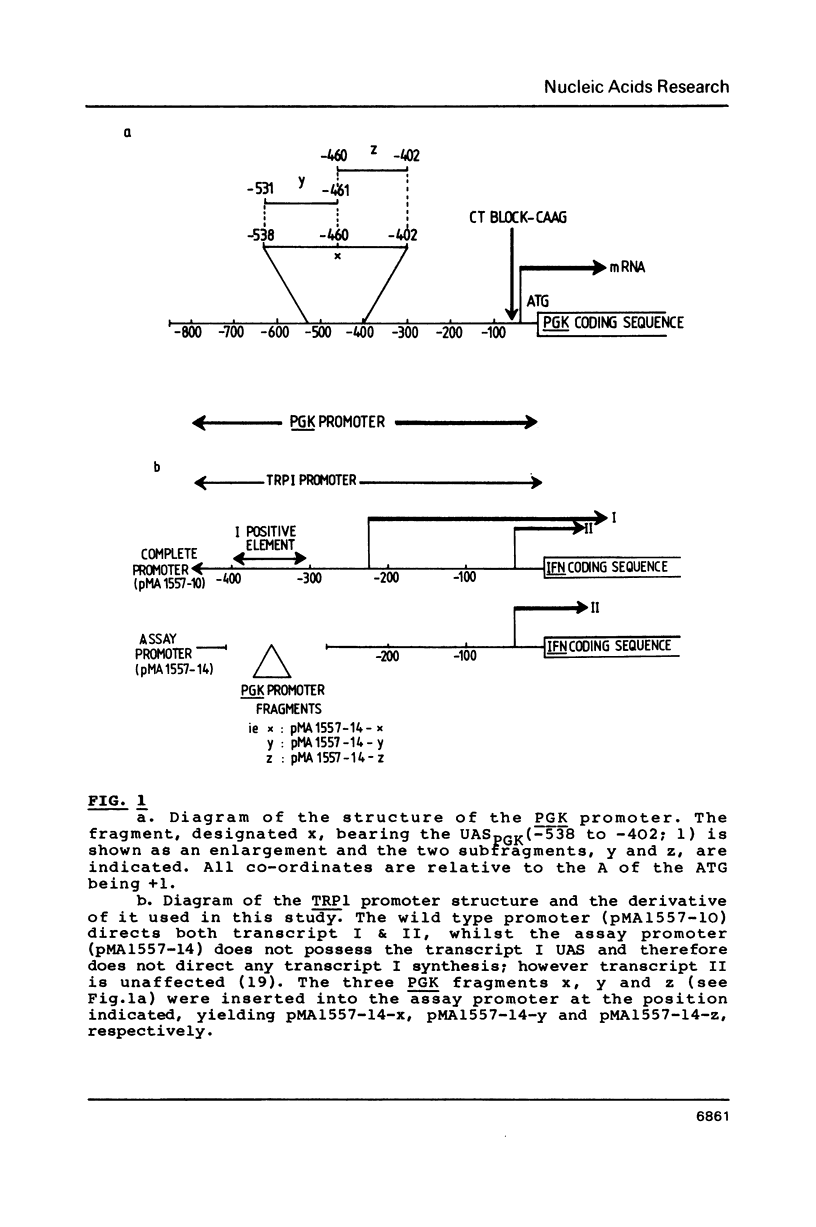
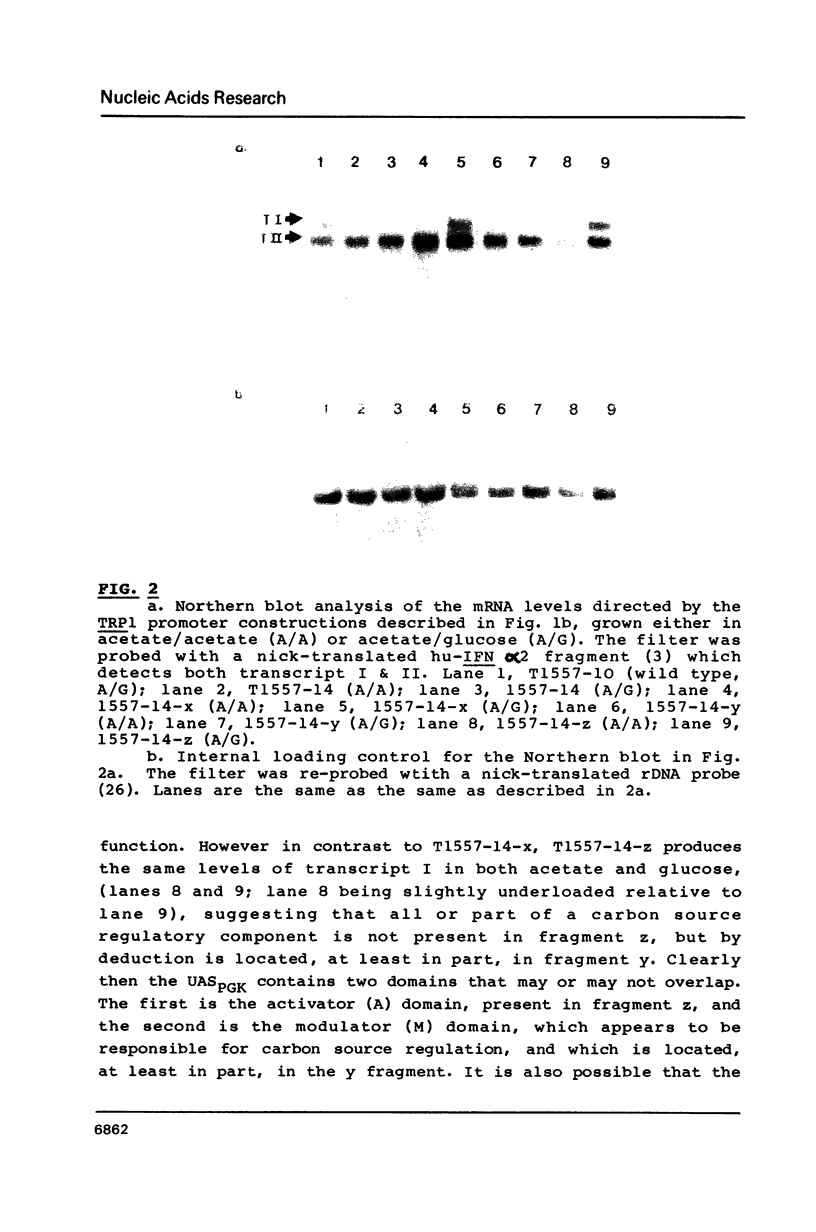
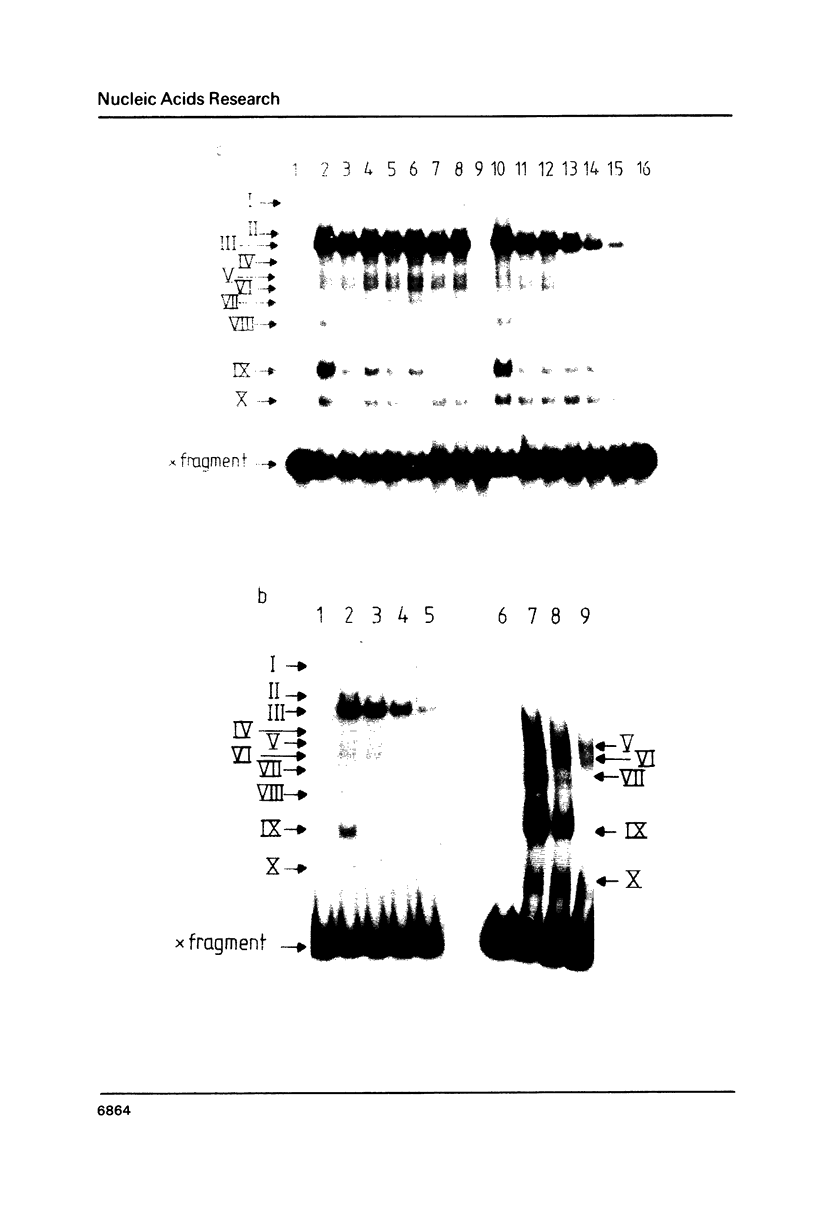
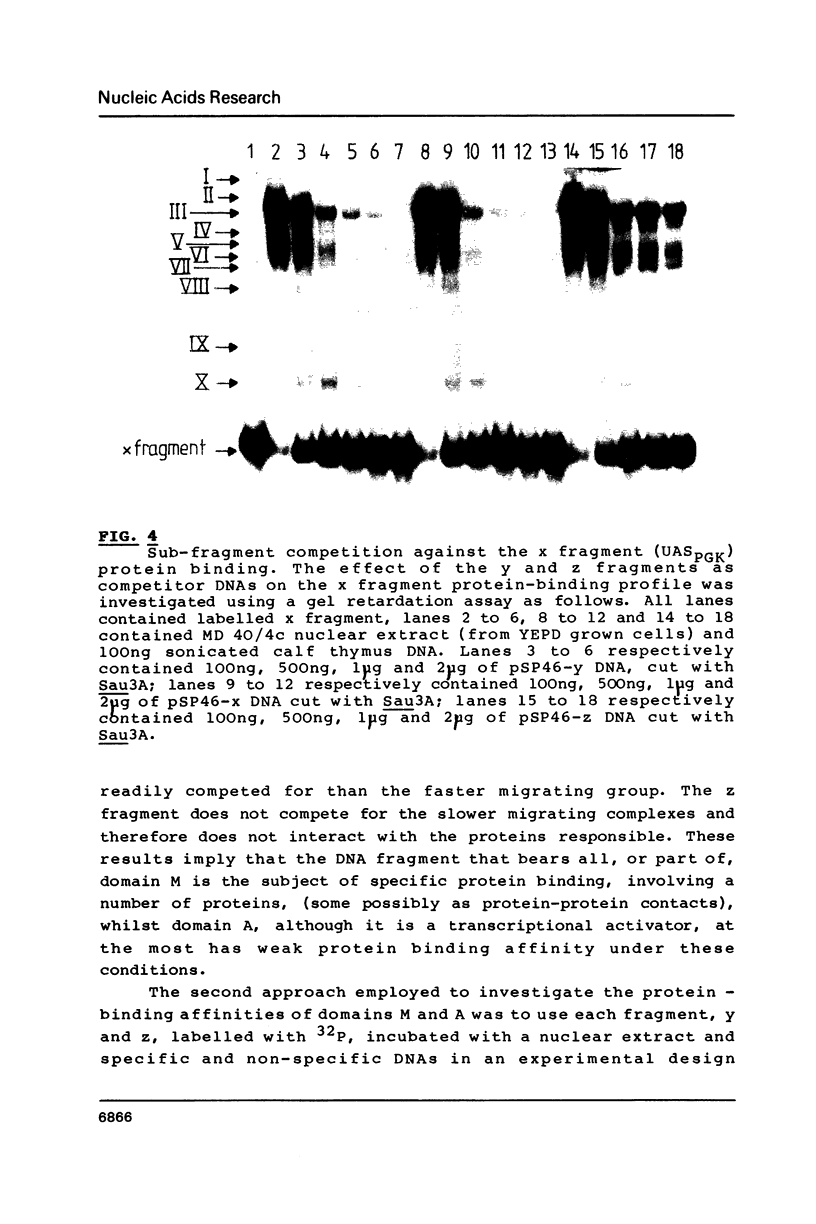
The upstream activation site (UAS) of the yeast phosphoglycerate kinase gene (PGK) has been localised by deletion analysis (1). Here we show that the UASPGK contains two functionally distinct domains. These two domains, designated activator (A) and modulator (M), appear to be located within bases -460 to -402 and -531 to -461, respectively, relative to the initiating ATG; although it is possible that part of the M domain resides within the A domain. They have been shown, using a heterologous assay promoter, to have distinct transcriptional functions. Domain A is responsible for activation of transcription whilst domain M is required for carbon source dependent regulation of transcription. Protein-DNA binding studies have demonstrated that the DNA fragment containing domain M has high affinity for at least one specific DNA-binding protein, whilst domain A does not appear to interact strongly in protein-binding assays under the same conditions. The domain M binding activity is dependent on the carbon source in the growth medium and may be functional in the carbon source control of PGK expression.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arcangioli B., Lescure B. Identification of proteins involved in the regulation of yeast iso- 1-cytochrome C expression by oxygen. EMBO J. 1985 Oct;4(10):2627–2633. doi: 10.1002/j.1460-2075.1985.tb03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Clifton D., Fraenkel D. G. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem. 1981 Dec 25;256(24):13074–13078. [PubMed] [Google Scholar]
- Cryer D. R., Eccleshall R., Marmur J. Isolation of yeast DNA. Methods Cell Biol. 1975;12:39–44. doi: 10.1016/s0091-679x(08)60950-4. [DOI] [PubMed] [Google Scholar]
- Dobson M. J., Mellor J., Fulton A. M., Roberts N. A., Bowen B. A., Kingsman S. M., Kingsman A. J. The identification and high level expression of a protein encoded by the yeast Ty element. EMBO J. 1984 May;3(5):1115–1119. doi: 10.1002/j.1460-2075.1984.tb01938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Hawthorne D C, Mortimer R K. Chromosome Mapping in Saccharomyces: Centromere-Linked Genes. Genetics. 1960 Aug;45(8):1085–1110. doi: 10.1093/genetics/45.8.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland M. J., Holland J. P. Isolation and identification of yeast messenger ribonucleic acids coding for enolase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate kinase. Biochemistry. 1978 Nov 14;17(23):4900–4907. doi: 10.1021/bi00616a007. [DOI] [PubMed] [Google Scholar]
- Hommes F. A. Effect of glucose on the level of glycolytic enzymes in yeast. Arch Biochem Biophys. 1966 Apr;114(1):231–233. doi: 10.1016/0003-9861(66)90325-0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Kim S., Mellor J., Kingsman A. J., Kingsman S. M. Multiple control elements in the TRP1 promoter of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Dec;6(12):4251–4258. doi: 10.1128/mcb.6.12.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsman S. M., Kingsman A. J., Dobson M. J., Mellor J., Roberts N. A. Heterologous gene expression in Saccharomyces cerevisiae. Biotechnol Genet Eng Rev. 1985;3:377–416. doi: 10.1080/02648725.1985.10647819. [DOI] [PubMed] [Google Scholar]
- Maitra P. K., Lobo Z. A kinetic study of glycolytic enzyme synthesis in yeast. J Biol Chem. 1971 Jan 25;246(2):475–488. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Roberts N. A., Kingsman A. J., Kingsman S. M. Factors affecting heterologous gene expression in Saccharomyces cerevisiae. Gene. 1985;33(2):215–226. doi: 10.1016/0378-1119(85)90096-4. [DOI] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Roberts N. A., Tuite M. F., Emtage J. S., White S., Lowe P. A., Patel T., Kingsman A. J., Kingsman S. M. Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene. 1983 Sep;24(1):1–14. doi: 10.1016/0378-1119(83)90126-9. [DOI] [PubMed] [Google Scholar]
- Nelson R. G., Fangman W. L. Nucleosome organization of the yeast 2-micrometer DNA plasmid: a eukaryotic minichromosome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6515–6519. doi: 10.1073/pnas.76.12.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden J. E., Stanway C., Kim S., Mellor J., Kingsman A. J., Kingsman S. M. Efficient expression of the Saccharomyces cerevisiae PGK gene depends on an upstream activation sequence but does not require TATA sequences. Mol Cell Biol. 1986 Dec;6(12):4335–4343. doi: 10.1128/mcb.6.12.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Hereford L. M., Skryabin K. G. Characterization of two types of yeast ribosomal DNA genes. J Bacteriol. 1978 Apr;134(1):295–305. doi: 10.1128/jb.134.1.295-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias P. P., DuBow M. S. The cloning and characterization of the bacteriophage D108 regulatory DNA-binding protein ner. EMBO J. 1985 Nov;4(11):3031–3037. doi: 10.1002/j.1460-2075.1985.tb04040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M. F., Dobson M. J., Roberts N. A., King R. M., Burke D. C., Kingsman S. M., Kingsman A. J. Regulated high efficiency expression of human interferon-alpha in Saccharomyces cerevisiae. EMBO J. 1982;1(5):603–608. doi: 10.1002/j.1460-2075.1982.tb01215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]