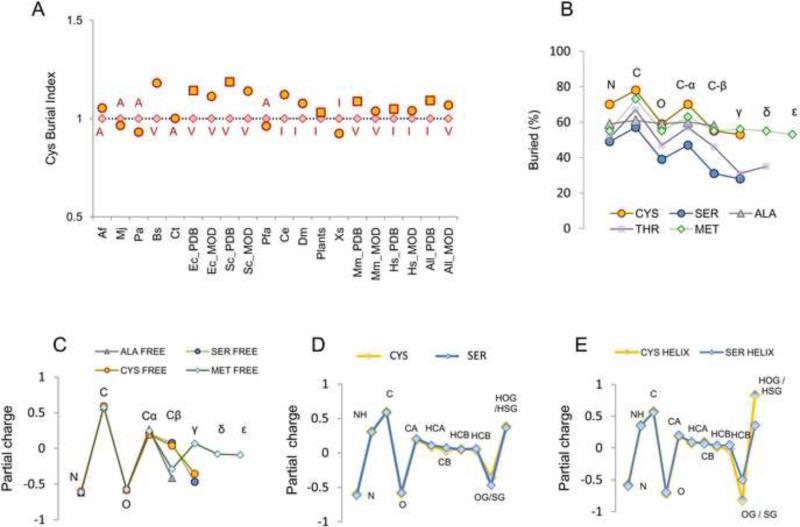
Figure 1. Cys is the least exposed residue in proteins, yet its chemical-physical properties are of a polar residue.
Cys exposure was calculated for proteins with known and modeled structures separated for each organism in the ModBase dataset, and for E. coli, S. cerevisiae and H. sapiens (the most represented organisms) in the PDB repository. Additionally, all non-redundant Viridiplantae PDBs, all PDB structures (“all PDB” column in panel A) and all ModBase models (all_MOD” column in panel A) were analyzed. Squared points highlight PDB structures, and circles ModBase structures. For each set of proteins, the proportion of completely buried (exposure for the whole residue <10 A2) normalized to the occurrence of this residue in the set was determined. To plot data for different organisms, we normalized percentage of buried Cys to the percentage of the most buried non-Cys residue within the same set (red rhombi refers to the most buried non-Cys residue, labeled in red in one letter code). This value is plotted in the Y-axis (labeled “Cys Burial Index”) in panel A. A value >1 (i.e., yellow circles, or yellow squares, higher than red rhombi) indicates that Cys is the most buried residue in the set. In the X-axis, abbreviations for organisms are reported, as defined in Methods. (B) Percentage of burial is shown for each composing atom of Cys, and for comparison, for each atom in Ser, Ala, Thr and Met, as calculated by the analysis of the PDB dataset. Above each point, the positions along the sidechain are reported (i.e., C-α, C-β, γ, δ, ε). To be noted, Thr is C-β branched (i.e., has two γ atoms and no δ atom): in the figure, for the sake of discussion (i.e., to visually compare it with Ser and Cys), only Oγ of Thr is aligned with γ atoms, while Cγ of Thr is aligned with Met δ atom. (C) Calculations with a QM approach are plotted as a function of atoms composing an amino acid residue. For comparison, we show residues with charge distribution similar to that of Cys (data provided by Dr. Annick Thomas and Dr. Robert Brasseur). (D) The same type of calculations for Cys and Ser residues outside defined secondary structures (i.e., not in the helix or strand), or belonging to α-helix (E) are shown. In panels D and E, to highlight differences between Ser and Cys in terms of the dipole on the functional group, hydrogen atoms are shown.