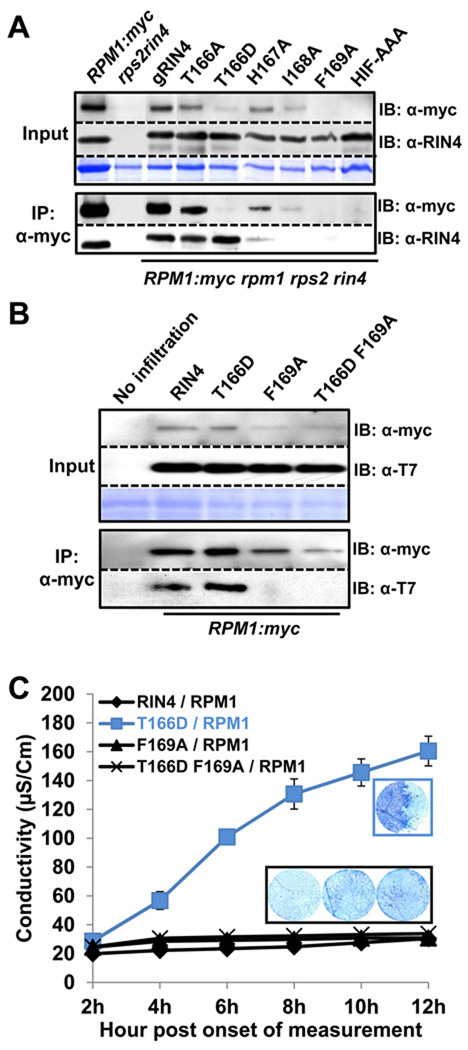
Figure 7. Differential co-immunoprecipitation of RPM1 with RIN4 BBS mutants identifies residues required for interaction and RPM1 accumulation.
(A) Co-immunoprecipitation of RIN4 BBS mutants with RPM1. The microsomal fraction was enriched in extracts from each RIN4 BBS mutant transgenic Arabidopsis, followed by immunoprecipitation with α-myc. The overall level of RPM1 is displayed in the input (left top). RIN4 expression in each mutant was confirmed by immunoblotting with α-RIN4. Immunoprecipitated RPM1 was shown by immunoblotting with α-myc. Co-immunoprecipitated RIN4 with RPM1 was confirmed with α-RIN4 immunoblot Two week-old seedlings from each line were used to collect the microsomal fraction.
(B) Loss of co-immunoprecipitation of RIN4 T166D F169A with RPM1. Agrobacterium transient assays were performed as in Figure 3. Loading controls, immunoprecipitation with α-myc and subsequent immunoblots were performed as in Figure 7A, with the use of α-T7 to detect RIN4 and RIN4 BBS mutants.
(C) Loss of effector-independent RPM1 activation in RIN4 T166D F169A. Agrobacterium transient assays, conductivity measurements and trypan blue staining were performed as in Figure 3.