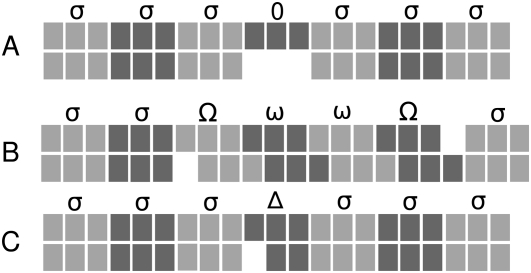
FIGURE 2.
Examples of typical gap patterns and scoring paths in a pairwise alignment assumed to be coding. Nucleotides are shown as blocks, codons as three consecutive blocks of the same shading. (A) A gap of length three does not change the reading frame and in-frame-aligned codons are scored with the normalized substitution score σ. (B) A single gap destroys the reading frame but gets corrected downstream by another gap. The triplets that are out-of-phase because of this obvious alignment error are penalized by the two frameshift penalties Ω and ω. (C) A single gap that, in principle, destroys the reading frame is interpreted as a sequence error. Penalized by a high negative score Δ, this frameshift is ignored, and downstream codons are considered to be in-phase.