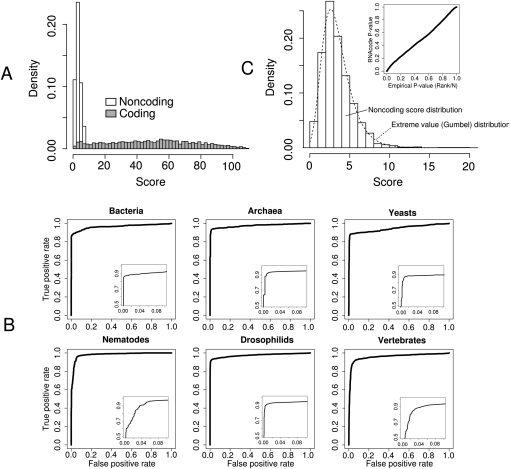
FIGURE 3.
RNAcode results on comparative test sets from various species. (A) Score distributions of annotated coding regions and randomly chosen noncoding regions in the Drosophila test set. (B) ROC curves for all six test sets. The full curve for all ranges of sensitivity/specificity from 0 to 1 is shown in the main diagrams. (Insets) The high specificity rate with false positive rates from 0 to 0.1. (C) Score distribution of noncoding alignments. The same distribution of the Drosophila test set as shown in A is shown in more detail. The fitted Gumbel distribution is shown as dotted line. (Upper right diagram) Comparison of the calculated P-values (via simulation and fitting of the Gumbel distribution) to the empirical P-values, i.e., the actual observed frequencies in the test set.