FIGURE 1.
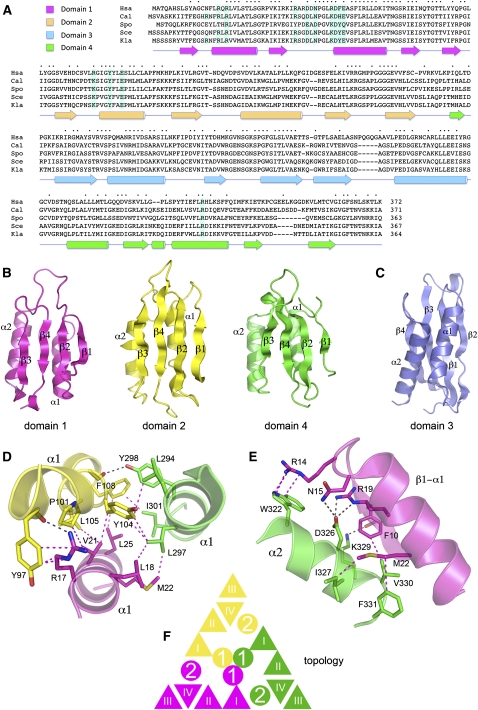
Rcl1 structure and domain organization. (A) The Rcl1 clade. The amino acid sequences of the Rcl1 proteins of Homo sapiens (Hsa), Candida albicans (Cal), Schizosaccharomyces pombe (Spo), Saccharomyces cerevisiae (Sce), and Kluyveromyces lactis (Kla) are aligned. Positions of amino acid side-chain identity/similarity are denoted by • above the sequences. Gaps in the alignment are indicated by –. The secondary structure elements of the KlaRcl1 fold (strands depicted as arrows and helices as cylinders) are shown below the sequence and are colored according to domain modules as specified at top, left (domain 1 in magenta, domain 2 in yellow, domain 3 in blue, and domain 4 in green). The conserved KlaRcl1 residues that were subjected to mutagenesis are highlighted in shaded boxes. (B) Rcl1 domain modules 1, 2, and 4 have a shared fold composed of a four-stranded β sheet plus two α helices arrayed with the same topology. (C) Rcl1 domain 3 has a distinctive folding topology. The secondary structure elements are labeled. (D) A three-helix bundle of the α1 helices of domains 1, 2, and 4 comprises the inner layer of the tri-radial domain architecture. Amino acid side chains and main-chain atoms engaged in cross-domain contacts are rendered as stick models. Hydrogen bonds are indicated by black dashed lines; van der Waals contacts are denoted by magenta dashed lines. (E) The α2 helices of domains 1, 2, and 4 comprise the middle tri-radial layer. The figure depicts the cross-domain contacts of the domain 4 α2 helix with the adjacent β1–α1 segment of domain 1. (F) Rcl1 domains 1, 2, and 4 are arrayed radially in three layers. The figure shows a cartoon diagram of the domain topologies. β strands are represented as triangles and α helices as circles.