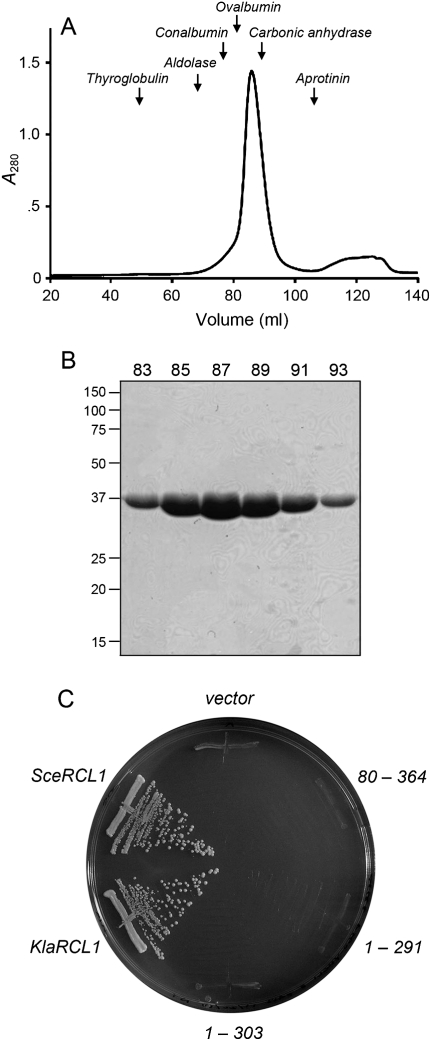
FIGURE 2.
Physical and genetic characterization of KlaRcl1. (A) Gel filtration. SeMet-KlaRcl1 (17.5 mg in 3.5 mL) was analyzed by Sephadex 200 gel filtration. The protein elution profile was monitored continuously by UV absorbance. A280 is plotted as a function of elution volume. The column was calibrated by tracking the elution profiles of marker proteins of known native size. The peak positions of the markers thyglobulin (669 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and aprotinin (6.5 kDa) are indicated by arrows. KlaRcl1 (a 39-kDa polypeptide) chromatographed as a single component eluting between ovalbumin and carbonic anhydrase. (B) Aliquots (5 μL) of the indicated Sephadex 200 column fractions were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker proteins are indicated on the left. (C) Complementation of S. cerevisiae rcl1Δ. Plasmid shuffle was performed with CEN HIS3 plasmids bearing wild-type SceRCL1, wild-type KlaRCL1, or the indicated KlaRCL1 deletion mutants. The empty CEN HIS3 plasmid vector served as a negative control. Single His+ transformants were streaked on agar medium containing 0.75 mg/mL of FOA; the plate was photographed after incubation for 3 d at 30°C.