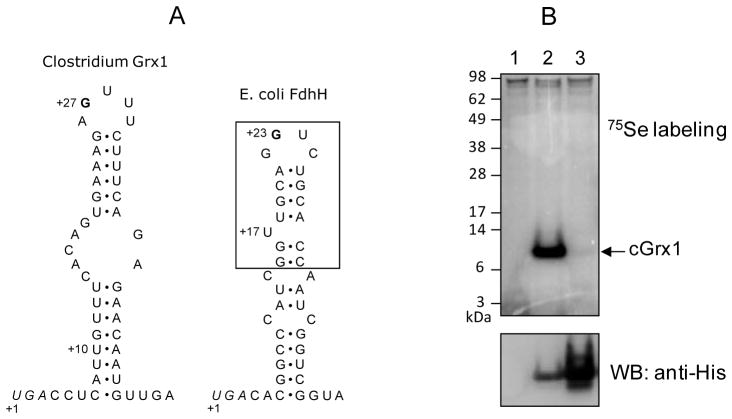
Figure 2. Structure of the SECIS element and expression of Clostridium selenoprotein Grx in E. coli.
(A) Structures of SECIS elements of Clostridium Grx1 (left) and E. coli fdhH (right). The Clostridium Grx1 SECIS contains a conserved G nucleotide in the apical loop, but lacks a bulged U, which is present in the minimal step-loop structure (boxed) in E. coli fdhH SECIS. Sec UGA codons are shown in italics and numbering of nucleotides is initiated at these Sec codons. The U at +10 position of Clostridium Grx1 SECIS is changed to A in monoselenol form of the protein. The SECIS structure of Clostridium Grx1 was predicted with RNAfold (Schuster et al., 1994). (B) 75Se metabolic labeling. E. coli BL21(DE3) cells transformed with an empty vector (lane 1), pET-CLOS-Grx1 (lane 2), or pET-CLOS-Grx1/U13C (lane 3) were metabolically labeled with 75Se. Proteins were separated by SDS-PAGE, transferred onto a PVDF membrane, and visualized with a PhosphorImager (upper panel). The same membrane was immunoblotted with anti-His antibody (lower panel).