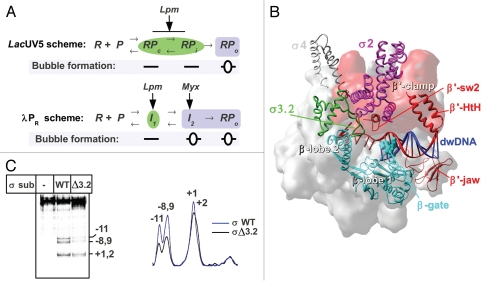
Figure 1.
(A) Kinetics scheme of RPo formation on the lacUV5,7 and λPR promoters.9 R: RNAP, P: promoter. Complexes with open DNA marked by blue boxes; closed complexes marked by green ellipses. (B) Structural model of RNAP in complex with dwDNA fragment.15 The structure of T. thermophilus RNAP12 is shown as a molecular surface colored in gray. The β lobes (in cyan), β' switch-2 (sw2) (Ser602-Lys621), β' jaw (Arg1266-Gly1328) and β' B-helix-turn-helix (HtH) (Leu469-Pro506)18 are shown as ribbons, the β-gate loop (Arg243-Pro248)12 is shown in CPK. Numbering corresponds to T. thermophilus. The β' clamp part of the RNAP surface is colored in red. DNA is shown in red (template) and blue (non-template). The σ subunit is shown as ribbons with the structural domains 2, 3 and 4 in magenta, green and gray, respectively. (C) Influence of the σ3.2HL deletion on lacUV5 promoter melting. KMnO4 probing of the open complexes formed between end-labeled lacUV5 promoter and the RNAP carrying either wild type σ70 or σ70 with deletion of the region 3.2 hairpin loop (Asp513-Leu519).3 Positions of the template strand thymines reactive to KMnO4 are indicated. The scan of the gel is shown on the right.