Abstract
There is growing interest in the use of cnidarians (corals, sea anemones, jellyfish and hydroids) to investigate the evolution of key aspects of animal development, such as the formation of the third germ layer (mesoderm), the nervous system and the generation of bilaterality. The recent sequencing of the Nematostella and Hydra genomes, and the establishment of methods for manipulating gene expression, have inspired new research efforts using cnidarians. Here, we present the main features of cnidarian models and their advantages for research, and summarize key recent findings using these models that have informed our understanding of the evolution of the developmental processes underlying metazoan body plan formation.
Keywords: Cnidaria, EvoDevo, Comparative genomics, Transgenics
Introduction
Cnidarians (corals, sea anemones, jellyfish and hydroids) form a diverse phylum that contains ~9000 species, which live in aquatic (predominantly marine) environments. The phylum-defining trait of Cnidaria is the stinging cell, the nematocyte (see Glossary, Box 1), an extrusive organelle used for predation, adhesion and defence (Holstein, 1981; Lengfeld et al., 2009; Tardent and Holstein, 1982). Cnidarians are divided into two major groups (Fig. 1): Anthozoa (sea anemones, corals and sea pens), which live as sessile polyps; and Medusozoa (jellyfish, sea wasps and Hydra), many, but not all, of which form a free-swimming medusa as well as polyps. Cnidarians in both groups have an external radial symmetry, yet internal asymmetries and bilaterality are displayed in many groups. Cnidarians have a single opening that acts as both mouth and anus and is generally surrounded by nematocyte-bearing tentacles.
Box 1. Glossary
- Autozooid.
A feeding polyp within a hydrozoan colony.
- Basal metazoans.
Early-branching animal lineages (excluding the Bilateria), i.e. Porifera (sponges), Ctenophora (comb jellies), Cnidaria (jellyfish) and Placozoa (Trichoplax).
- Bilateria.
Bilaterally symmetrical animals.
- Blastopore.
The site of gastrulation, i.e. of the formation of inner germ layers, endoderm and mesoderm.
- Colonial thecate hydrozoan.
A hydrozoan colony that originates from a single primary polyp that either branches in a regular pattern (like branches on trees) or grows stolons or mats at the bases of polyps from which other polyps form, often with distinct functions. Thecate hydrozoans form an exoskeleton (theca) around the polyps and stolon mat; by contrast, athecate hydrozoans lack this structure.
- Coral bleaching.
Whitening, caused by loss of symbiont dinoflagellates, that leads to the death of corals.
- Dinoflagellates.
A group of unicellular algae, some species of which are symbionts of corals and anemones.
- Dioecious.
Separate sexes.
- Ectoderm.
The outer tissue layer of a cnidarian polyp, larva or medusa.
- Endoderm.
The inner tissue layer of a cnidarian polyp, larva or medusa.
- Endomesoderm.
A germ layer in animal embryos that can give rise to either endoderm or mesoderm.
- Gonozooid.
A specialized polyp within a hydrozoan colony that forms only medusae or gametes.
- Hermaphrodite.
An animal form that has both male and female reproductive organs.
- Hox code.
Staggered expression of Hox genes along the anterior-posterior body axis in Bilateria that specifies axial identities and is colinear with Hox gene organization in the genome.
- Mesentery.
Endodermal folds in anthozoan polyps that extend from the body wall into the gastric cavity. In the oral part, the mesenteries are connected to the endodermal part of the pharynx.
- Mesoderm.
The third germ layer, situated between ectoderm and endoderm in Bilateria, which gives rise to many derivatives, such as blood, muscles and bones.
- Mesogloea.
Extracellular matrix between ectoderm and endoderm in cnidarian polyps and medusae; it is composed of laminins, collagens and proteoglycans.
- Nematoblasts.
Nematocyte precursors that exist in proliferating clusters of cells connected by cytoplasmic bridges.
- Nematocyte.
A cell type, unique to cnidarians, that contains an extrusive organelle (the nematocyst) used for prey capture, defence or adhesion. Hydra, for example, has four types of nematocytes: stenoteles, desmonemes, atrichous isorhizas and holotrichous isorhizas.
- Planula larva.
The typical ciliated postgastrula larval stage in cnidarians.
- Reaction-diffusion-based mechanism.
A mechanism for generating patterns, based on autocatalytic local activation and cross-activated long-range lateral inhibition; first formalized by Alan Turing, further developed by Alfred Gierer and Hans Meinhardt, particularly as applied to patterning in Hydra (Gierer and Meinhardt, 1972).
- Scleractinian corals.
Stony corals (Hexacorallia; e.g. Acropora millepora).
- Siphonoglyph.
Ciliated rim on one or both ends of the slit-like pharynx, defining the symmetry of the directive axis.
- Spemann-Mangold organizer.
The dorsal lip of the blastopore of the amphibian gastrula stage embryo, which is able to induce a secondary body axis upon transplantation to the ventral side of a host embryo.
- Stolon.
A tube extending laterally from the aboral end of a polyp in a colonial hydrozoan. As the stolon grows, new polyps arise from it in a spaced manner.
- Triploblastic.
Animals (including all bilaterians) with three germ layers: ectoderm, endoderm and mesoderm; in contrast to diploblastic animals (e.g. Cnidaria) with two germ layers (ectoderm and endoderm).
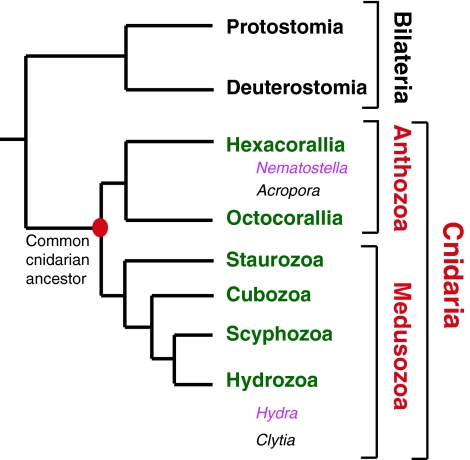
Fig. 1.
Phylogenetic relationships of classes in the phylum Cnidaria. A phylogenetic tree [based on the results of Collins (Collins, 2002) and Collins et al. (Collins et al., 2006)] showing the relationships within the phylum Cnidaria. The two main divisions of Cnidaria (Anthozoa and Medusozoa) are indicated in red. Anthozoa is a class that contains two subclasses (green), whereas Medusozoa is a subphylum consisting of four classes (green). Sequenced genomes (pink) are available for Nematostella and Hydra (Chapman et al., 2010; Putnam et al., 2007), whereas the genomes of Acropora and Clytia are currently being sequenced (black).
In recent years, interest in the use of cnidarians and other basal metazoans (see Glossary, Box 1) as model organisms has grown, particularly with the goal of understanding the evolution of the Bilateria (see Glossary, Box 1). Cnidaria is an early-branching metazoan lineage, and four of the five current cnidarian classes (Anthozoa, Hydrozoa, Cubozoa, Scyphozoa and Staurozoa) have been identified in the Cambrian fossil record (Cartwright et al., 2007), indicating that considerable cnidarian diversification had occurred by ~500 million years ago. Cnidaria, in phylogenetic terms, is a sister group to Bilateria (Fig. 1). Thus, questions regarding the evolution of key bilaterian traits can be profitably explored using cnidarians.
The main body plan features that distinguish Bilateria from Cnidaria are the bilaterality of the body axes, the presence of a third germ layer (mesoderm; see Glossary, Box 1) and a centralized nervous system. Cnidarians are generally considered to have only two germ layers (endoderm and ectoderm; see Glossary, Box 1) and are radially symmetric, although anthozoan polyps display an internal bilaterality in the asymmetric organization of the pharynx, the siphonoglyph (see Glossary, Box 1), and the retractor muscles in the mesenteries (see Glossary, Box 1) (e.g. Faurot, 1895; Faurot, 1903; Hyman, 1940; Jägersten, 1955) (for a review, see Salvini-Plawen, 1978). How this anthozoan asymmetry and the oral-aboral axis of polyps and medusae relate to the two body axes of bilaterians has been a long-standing question that has evoked numerous hypotheses and fuelled recent research. In this Primer, we provide an overview of current model cnidarians and highlight how recent findings using these models have advanced our understanding of bilaterian evolution and development. We also discuss the challenges that researchers need to overcome to expand developmental studies using cnidarians.
Model cnidarians: their habitats and life cycles
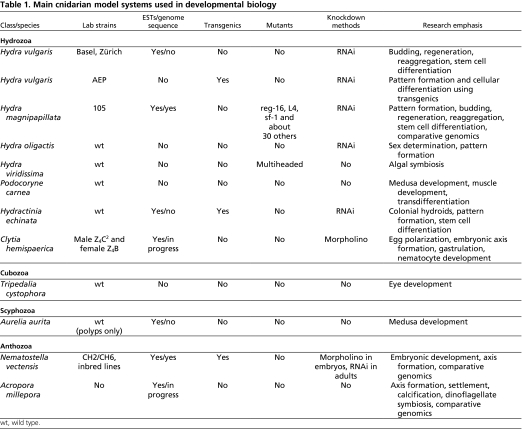
There are two main types of cnidarian life cycle. In anthozoans, the polyp is the gamete-producing form and the cycle is embryo>larva>polyp. Medusozoans generally have an embryo>larva>polyp>medusa life cycle, in which the medusa is typically the sexual form (Fig. 2). However, considerable variation exists within these general schemes. Below, we discuss the life cycles and habitats of the four model cnidarians used most widely for molecular studies of development and with completed or ongoing genome sequencing projects. Additional cnidarian species used for developmental studies are included in Table 1.
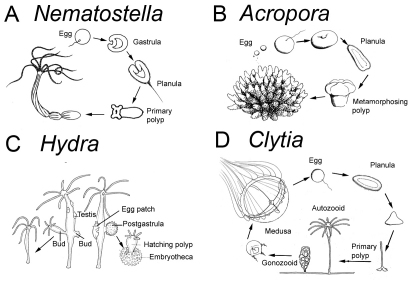
Fig. 2.
Life cycles of the main cnidarian model systems. (A,B) Anthozoan polyps either burrow into the soft substrate (A), here exemplified by the edwardsiid sea anemone Nematostella vectensis, or are attached to the surface (B), as with many sea anemones and corals, here exemplified by Acropora millepora. Nematostella female polyps release packages of several hundred eggs into the water, where they are fertilized (Fritzenwanker and Technau, 2002; Hand and Uhlinger, 1992). The resulting embryos develop into ciliated planula larvae that undergo either a gradual (Nematostella) or more dramatic (Acropora) metamorphosis into a sessile primary polyp, which involves calcification and formation of the skeleton in corals. (C) In Hydra, gametes develop from interstitial stem cells located in the ectoderm that differentiate within several testes or within a single egg patch. The embryo remains attached to the parent polyp from fertilization through gastrulation. The postgastrula embryo forms a cuticle from which the primary polyp hatches after several weeks or months. (D) The hydrozoan Clytia hemisphaerica forms a colony with feeding polyps (autozooids) and medusae-bearing gonozooids. Gametes are released from the medusae into the water. The embryo develops into a planula larva that settles to transform into a primary polyp, which then forms a new colony. Drawings are by Hanna Kraus (A,B,D) or modified from Tardent (Tardent, 1978) with permission (C).
Table 1.
Main cnidarian model systems used in developmental biology
Nematostella vectensis (Anthozoa)
During the last decade, Nematostella vectensis has become a leading model cnidarian system and was the first cnidarian to have its genome sequenced (Putnam et al., 2007). It is a brackish water sea anemone found on the American Atlantic and Pacific coasts and on England's south coast (Darling et al., 2004; Darling et al., 2005; Pearson et al., 2002; Sheader et al., 1997). Nematostella has a typical anthozoan life cycle (Fig. 2A, Fig. 3A-C). After about a week, the planula (see Glossary, Box 1) settles and metamorphoses into a polyp, which reaches sexual maturity in 3-6 months (Fritzenwanker and Technau, 2002; Hand and Uhlinger, 1992). The ease of culturing and maintaining Nematostella in the laboratory (Fritzenwanker and Technau, 2002; Genikhovich and Technau, 2009) has greatly facilitated its use as a developmental system.
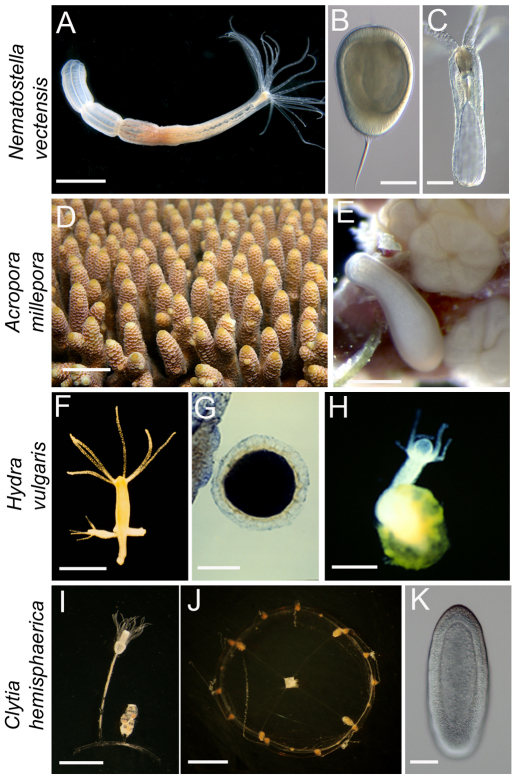
Fig. 3.
Cnidarian model systems used in developmental biology. (A-C) Nematostella vectensis, showing adult polyp (A), planula larva (B) and primary polyp (C). (D,E) Acropora millepora showing coral (D) and planula larva and metamorphosing early settlement stages (E). (F-H) Hydra vulgaris showing budding polyp (F), cuticle stage postgastrula embryo (G) and hatching primary polyp (H). (I-K) Clytia hemisphaerica showing autozooid and gonozooid polyps (I), young medusa (J) and planula larva (K). Note the differences in size between different cnidarians. All polyps and planulae are oriented with oral side up (except for A). Images were taken by Jens Fritzenwanker and U.T. (A-C), David Miller (D), Eldon Ball (E), Tim Nüchter and Thomas Holstein (F), U.T. (G,H) and Hanna Kraus and U.T. (I-K). The images in B and C are reproduced with permission (Rentzsch et al., 2008). Scale bars: 1 cm in A; 70 μm in B,K; 80 μm in C; 5 cm in D; 150 μm in E; 500 μm in F,J; 250 μm in G,H; 100 μm in I.
Acropora millepora (Anthozoa)
Scleractinian corals (see Glossary, Box 1) of the genus Acropora are major contributors to the Australian Great Barrier Reef and to other pacific reefs. Acropora millepora has been used as a model for coral development, and, like other anthozoans, has polyp, embryo and planula larva stages (Fig. 2B, Fig. 3D,E). Like other corals, A. millepora cannot currently be kept in the laboratory through a full life cycle. In the wild, the planula larva stage may persist for months. It spawns for only 1-2 days in spring, which limits the availability of material and restricts experimental studies of its development to the examination of gene expression patterns. Nevertheless, important evolutionary insights have been obtained from such expression studies and from analyses of A. millepora expressed sequence tag (EST) datasets (Ball et al., 2002; Galliot and Miller, 2000). Studies of A. millepora are also important for comparison with other anthozoans (e.g. sea anemones) and for studying the formation of the calcified skeleton that defines the coral reef. A. millepora also contains intracellular symbiotic dinoflagellates (see Glossary, Box 1), and so is an important system for understanding how this symbiosis is established and maintained, which has a bearing on the current widespread phenomenon of coral bleaching (see Glossary, Box 1).
Hydra (Hydrozoa)
Hydra is a freshwater polyp with a cosmopolitan distribution that never forms a medusa and is considered to be the first model system to have been used for experimental developmental biology (Trembley, 1744). It reproduces primarily asexually by budding of lateral polyps, but it also has a sexual cycle (Fig. 2C, Fig. 3F-H). Polyps are either hermaphroditic or dioecious (see Glossary, Box 1), depending on the strain or species. Well-fed polyps produce a new bud in 3-4 days. The conditions that induce the formation of sperm and eggs are not entirely understood, but involve temperature in some species (e.g. H. oligactis) or starvation (H. vulgaris). There is no larval stage between embryo and polyp in Hydra. The embryo completes development within a cuticle from which a fully formed polyp hatches after a dormancy phase of weeks to months (Fig. 2C, Fig. 3H) (Bottger et al., 2006; Brauer, 1891).
The main Hydra models are H. vulgaris and H. magnipapillata. However, these are unlikely to be separate species; H. magnipapillata should probably be renamed H. vulgaris (Martinez et al., 2010). Strain 105 of H. magnipapillata was used for the Hydra genome project (Chapman et al., 2010). Strain AEP of H. vulgaris, which is the result of a cross of two North American isolates, is hermaphroditic and continuously produces gametes and embryos with an average dormant period of only 4 weeks in the laboratory (Bottger et al., 2006). This strain has been used to study gametogenesis and embryogenesis (Martin et al., 1997; Miller et al., 2000; Technau et al., 2003; Wittlieb et al., 2006) and, most importantly, is the source of embryos for making transgenic Hydra (Wittlieb et al., 2006).
Clytia hemisphaerica (Hydrozoa)
Clytia is a cosmopolitan colonial thecate hydrozoan (see Glossary, Box 1). Unlike Hydra, it has a typical hydrozoan life cycle that includes planula larva and medusa stages (Fig. 2D, Fig. 3I-K). The colony contains two types of polyps: gonozooids and autozooids (see Glossary, Box 1 and Fig. 2D). Clytia is an important new cnidarian model that offers many advantages owing to its total transparency, its ease of culturing both sexually and asexually in the laboratory, and its tractability as an experimental system (for a review, see Houliston et al., 2010). Clytia has been used to study the role of Wnt signaling in egg polarization and in nematocyte differentiation in the medusa (Denker et al., 2008; Momose et al., 2008; Momose and Houliston, 2007). A large Clytia EST dataset is publicly available and sequencing of the Clytia genome is in progress (Houliston et al., 2010).
The cnidarian body plan and its regeneration
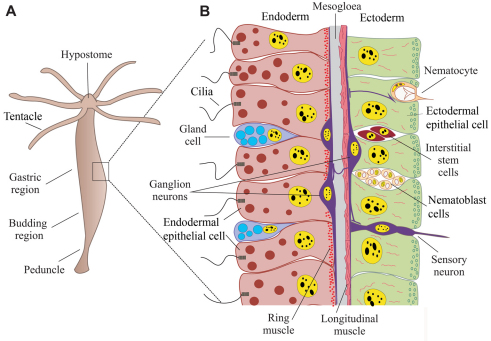
Cnidarians have one body axis and only two cell layers, ectoderm and endoderm, which are separated by an extracellular matrix called the mesogloea (Fig. 4). Including the various nematocyte subtypes, a typical cnidarian has around a dozen morphologically distinguishable cell types, as shown in Fig. 4B for Hydra. However, at the molecular level, greater cell type diversity exists in cnidarians than is indicated by morphology alone. For example, morphologically indistinguishable nerve cells can have very different patterns of gene expression (Bode, 1992; Galliot et al., 2009; Technau and Holstein, 1996).
Fig. 4.
Anatomy of a hydrozoan polyp. (A) A Hydra polyp is essentially a two-layered tube, with a ring of tentacles around the mouth opening at the tip of the hypostome. Asexual budding occurs on the lower half of the body column. Interstitial stem cells and nematoblasts are distributed evenly in the body column, below the tentacle ring and above the border of the peduncle, which is the stalk between the budding region and pedal disc. (B) The bilayered cellular organization of a Hydra polyp. Ectoderm and endoderm are separated by an acellular matrix called the mesogloea (gray). All epithelial cells in Hydra are myoepithelial, with myofibers on the basal side (red). In ectodermal epithelial cells (green), the fibers are oriented longitudinally, and in endodermal epithelial cells (pink) they are oriented circumferentially (ring muscle). Most interstitial cells and nematoblast clusters are located between ectodermal epithelial cells. Neurons are found in both the endoderm and ectoderm. Sensory neurons are located between epithelial cells and connect to ganglion neurons (purple), which are at the base of the epithelium on top of the myofibers and sometimes cross the mesogloea. Different types of gland cells, most of which are found in the endoderm, are intermingled between the epithelial cells.
Many cnidarians are able to propagate asexually, for example by budding in Hydra. Such asexual propagation requires the continuous production of differentiated cell types. In hydrozoans, asexual propagation has been shown to involve the production of new cells by division and subsequent differentiation. In Hydra, which is the best-studied case, new epithelial cells arise by the division of existing epithelial cells, whereas neurons, nematocytes, gland cells and gametes arise from multipotent stem cells called interstitial cells (David and Murphy, 1977) (reviewed by Bode, 1996; Bosch, 2009). Interestingly, interstitial stem cells have so far not been reported outside of the class Hydrozoa and it is unclear how differentiated cell types arise during asexual propagation in these other classes of cnidarians.
A striking feature of cnidarians is their ability to regenerate. For example, a Hydra polyp, when bisected, will regenerate the missing oral and aboral structures within 2-4 days (Trembley, 1744). In fact, Hydra can be cut into ~20 fragments, each of which will regenerate a complete polyp (Bode and Bode, 1980). Even more extraordinarily, Hydra can be dissociated into a suspension of cells which, when reaggregated, regenerates polyps de novo, consistent with a reaction-diffusion-based mechanism (see Glossary, Box 1) of pattern formation (Gierer et al., 1972; Technau et al., 2000; Technau and Holstein, 1992). Regeneration in Hydra is a morphallactic process, i.e. it occurs by a mechanism that does not require proliferation and growth (Cummings and Bode, 1984). Considerable effort is now being made to explain regeneration in Hydra at the molecular level, by examining the involvement of conserved signaling pathways and transcription factors, such as the Wnt pathway and Brachyury (Bielen et al., 2007; Broun and Bode, 2002; Broun et al., 2005; Chera et al., 2009; Galliot, 2000; Galliot et al., 2008; Gee et al., 2010; Hobmayer et al., 2000; Holstein et al., 1991; Holstein et al., 2003; Lengfeld et al., 2009; Technau and Bode, 1999). The molecular basis of regeneration in other cnidarians is also beginning to be explored at the molecular level (Burton and Finnerty, 2009).
Experimental techniques available in cnidarians
The toolkit available for experimental studies of cnidarian development is substantial and has expanded particularly rapidly with the advent of molecular and genomic technologies. We provide a brief survey of the major molecular genetic approaches below and a summary of methods for manipulating and analyzing tissues and embryos in Box 2.
Box 2. Experimental analysis in cnidarians
Manipulations of tissues and cells in polyps and embryos
Transplantation experiments (Browne, 1909), in which a piece of tissue was grafted laterally onto a host polyp, revealed the involvement of two developmental gradients in Hydra head formation: a head activation gradient, i.e. the capacity of the graft to induce secondary head formation dependent on its axial origin; and the head inhibition gradient, i.e. the gradient of suppression of secondary head formation dependent on the distance from the host head (MacWilliams, 1983a; MacWilliams, 1983b). Numerous variations on these classical transplantations have been carried out (MacWilliams, 1983a, MacWilliams, 1983b; Broun and Bode, 2002; Gee et al., 2010), including recombination of tissue layers (Schmid and Tardent, 1984; Takano and Sugiyama, 1984), removal and transplantation of interstitial cells (Campbell, 1976; Heimfeld and Bode, 1984) and the dissociation and reaggregation of Hydra cells (Gierer et al., 1972; Technau et al., 2000). Grafting, dissociation and separation of blastomeres and lineage tracing have also been successfully performed in cnidarian embryos (Freeman, 1990; Fritzenwanker et al., 2007; Kraus et al., 2007; Lee et al., 2007; Momose and Schmid, 2006).
Cell cycle analysis
Labeling of S-phase cells with bromodeoxyuridine or 3H-thymidine has been used to study proliferation in cnidarians and has revealed that, in at least some cnidarians, cells apparently lack a G1 phase (Campbell and David, 1974; David and Campbell, 1972; David and Gierer, 1974; Plickert et al., 1988).
Use of small molecules to study cnidarian development
Cnidarian polyps and embryos are permeable to small molecules that perturb signaling pathways. The GSK3β inhibitor alsterpaullone was used to show that the head organizer in Hydra functions through the canonical Wnt pathway (Broun et al., 2005). The Wnt pathway has also been manipulated by diacylglycerol and LiCl (Hassel and Bieller, 1996; Muller, 1990). Furthermore, chemical inhibitors of the fibroblast growth factor (FGF) receptor (SU5421) and of the mitogen-activated protein kinase kinase MEK (UO129) have been successfully applied during Nematostella embryonic and larval development (Rentzsch et al., 2008).
Methods for gene manipulation in cnidarians
mRNA injection
Gain-of-function studies by injection of synthetic mRNAs and loss-of-function studies by injection of mRNAs encoding dominant-negative versions of proteins have been performed successfully in embryos from Nematostella and Clytia (Lee et al., 2007; Momose and Houliston, 2007; Wikramanayake et al., 2003). However, owing to the short lifetime of the mRNA, only early embryonic stages can be assessed.
RNAi
The first use of RNA interference (RNAi) in a cnidarian was reported in 1999 (Lohmann et al., 1999), when double-stranded (ds) RNA was introduced into adult Hydra polyps by electroporation. Later, localized electroporation of dsRNA, using a pipette applied to early buds in Hydra, was performed to locally knockdown endogenous transcripts (Smith et al., 2000). As a proof of the specificity of RNAi in Hydra, it has been shown that transgenic green fluorescent protein (GFP) can be knocked down efficiently by electroporation of dsRNA without any other phenotype (Khalturin et al., 2008). More recently, the RNAi feeding method from C. elegans (Conte and Mello, 2003) has been adapted to Hydra by introducing agarose particles that contain dsRNA-producing bacteria into the gastric cavity of the adult polyp (Miljkovic-Licina et al., 2007). However, the application of RNAi to Hydra has its limitations. To be effective, electroporation in a cuvette leads to greater than 50% mortality during the first day, even in controls. The bacteria feeding approach requires treatment for up to 18 days and ultimately leads to starvation and cell death. Thus, RNAi in Hydra is not yet a robust technology. RNAi has been used in studies of cnidarians in addition to Hydra (Duffy et al., 2010; Dunn et al., 2007; Kunzel et al., 2010; Pankow and Bamberger, 2007; Sunagawa et al., 2009). As such studies progress, one might expect RNAi to become a reliable tool for investigating gene function in cnidarian developmental models.
Morpholinos
Morpholino-mediated knockdown of endogenous transcripts has been successfully performed in Nematostella and Clytia (Momose et al., 2008; Momose and Houliston, 2007; Rentzsch et al., 2008; Saina et al., 2009). In these two cnidarians, microinjection of embryos is easy to carry out, making the delivery of morpholinos straightforward. However, owing to the necessity of injecting embryos or eggs, morpholino-mediated gene knockdown may only work for early embryonic stages and is not a suitable approach for embryos such as those of Hydra, which enter a dormant cuticle stage of variable length after gastrulation.
Transgenics
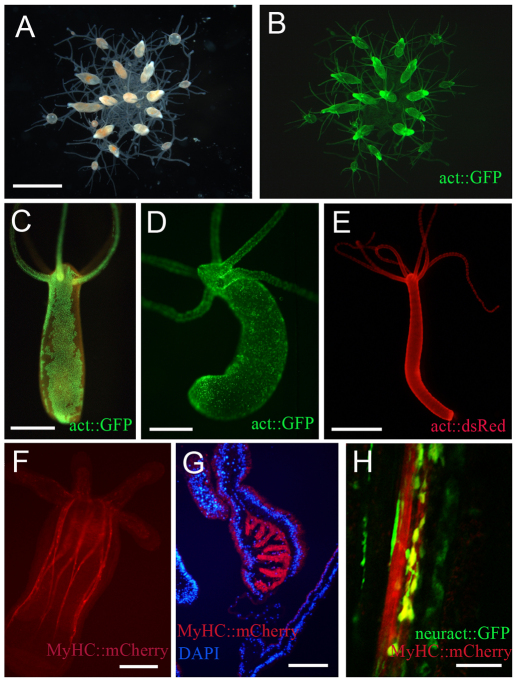
Efforts to introduce transgenes into Hydra were initiated in the late 1980s. Ultimately, transient expression of GFP under the control of a Hydra actin promoter was achieved by bombardment of adult Hydra polyps with gold particles coated with plasmid DNA (Bottger et al., 2002). Although this approach does not yield stably integrated transgenes, it has been useful for examining the localization of proteins in the transiently transgenic cells (Bottger et al., 2002; Kasbauer et al., 2007). A major breakthrough in research on cnidarian development came with the generation of stably transgenic Hydra by the injection of plasmid DNA into the embryo (Wittlieb et al., 2006) (Fig. 5). This was followed by the successful production of transgenic Nematostella that achieved germline transmission of an mCherry transgene, expressed from the myosin heavy chain promoter in the retractor muscles (Renfer et al., 2010) (Fig. 5). Recently Hydractinia has joined the club of transgenic cnidarians (Kunzel et al., 2010) (Fig. 5). The relative ease with which transgenesis has been achieved for these three diverse cnidarians suggests that it should be possible to generate transgenic animals from additional cnidarian species. Initial studies with transgenic Hydra and Hydractinia have provided new insights into patterning and stem cell biology in cnidarians (Khalturin et al., 2007; Kunzel et al., 2010; Siebert et al., 2005; Wittlieb et al., 2006), indicating the value of this approach.
Fig. 5.
Transgenic cnidarians. (A,B) A transgenic colony of the marine hydrozoan Hydractinia echinata, driving enhanced green fluorescent protein (eGFP, green) under the control of an actin promoter (act::GFP) in all cells. Dark-field (A) and fluorescent (B) images are shown. (C-E) Transgenic Hydra, with the oral end up. (C) A somatic patch of transgenic ectodermal epithelial cells expressing eGFP under the control of an actin promoter, demonstrating normal axial tissue displacement with growth. (D) Somatic first generation transgenic line expressing act::GFP only in the interstitial cell lineage and its derivatives as a result of late integration after segregation of the stem cell lineage. (E) Transgenic Hydra expressing an actin promoter-driven DsRed2 transgene (act::dsRed, red) in the ectoderm. (F-H) Transgenic Nematostella. (F) Transgenic F1 primary polyp expressing mCherry (red) under the control of a muscle-specific promoter (MyHC::mCherry). (G) Cryo-cross section through the mesentery of an adult polyp showing retractor muscle-specific transgene expression (red) and nuclei staining (DAPI, blue). (H) Confocal longitudinal section of a mesentery of a double-transgenic line expressing a neuron-specific transgene (neuract::GFP, green) and a marker of transgenic retractor muscles (MyHC::mCherry, red) showing close association (merge in yellow) of neurons with muscle cells. Images courtesy of Günter Plickert (A,B), Thomas C. Bosch (C,D), Catherine Dana and R.E.S. (E) and E. Renfer and U.T. (G,H). Image in F reproduced with permission (Renfer et al., 2010). Scale bars: 2 mm in A; 500 μm in C-E; 200 μm in F; 250 μm in G; 100 μm in H.
Key recent findings and their impact
When molecular studies of cnidarians were initiated, a major goal was to determine whether the genetic toolkit used to construct the bilaterian embryo (represented primarily by the model systems Drosophila, C. elegans, amphibians, zebrafish and mice) was in place in the ancestor of cnidarians and bilaterians. As we discuss below, recent research on cnidarians using molecular methods has helped to address this and other important questions in developmental biology.
How does the genetic toolkit that is involved in the development of the morphologically simple cnidarians compare to that used in bilaterians?
Despite their relatively simple anatomies, cnidarians have a surprisingly complex genetic toolkit. The first evidence for this came from EST datasets from Acropora and Nematostella (Ball et al., 2004; Kortschak et al., 2003; Miller et al., 2005; Technau et al., 2005), and sequencing of the Nematostella genome corroborated this view (Miller and Ball, 2008; Putnam et al., 2007). A striking example of the complexity of the cnidarian gene set comes from the finding that Nematostella has all of the Wnt family members (except for the Wnt9 subfamily) found in bilaterians (Kusserow et al., 2005; Lee et al., 2006). Comparison of the Nematostella and Hydra genome sequences (browsers are available for both genomes: Hydra, http://hydrazome.metazome.net/cgi-bin/gbrowse/hydra; Nematostella, http://www.metazome.net/cgi-bin/gbrowse/Nvectensis) shows that Hydra has undergone considerable gene loss compared with Nematostella (Chapman et al., 2010). Other recent studies have revealed the evolution and expansion of taxon-restricted genes in cnidarians, i.e. genes that have no counterpart in other lineages and which therefore might be involved in the evolution of lineage-specific morphological traits, such as nematocytes (Foret et al., 2010; Hwang et al., 2010; Khalturin et al., 2008; Khalturin et al., 2009; Steele and Dana, 2009; Steele et al., 2011).
How do the axes of cnidarians relate to the anterior-posterior and dorsal-ventral axes of bilaterians?
The anterior-posterior (A-P) axis in bilaterians (experimentally best exemplified in mice and flies) is specified by the combinatorial action of Hox genes that are expressed in a staggered manner (the ‘Hox code’; see Glossary, Box 1) along the axis, colinear with their clustered organization in the genome. The presence of a Hox cluster and colinear expression is taken as an indication of a conserved role for Hox genes in A-P body axis specification. Current findings indicate that the evolutionary history of Hox (and ParaHox) genes in cnidarians is complex (involving, for example, secondary losses and dramatically variable expression patterns) and that the history of genome organization for these genes is difficult to reconstruct, particularly as it relates to the Hox and ParaHox clusters in bilaterians. It seems likely, however, that a Hox code does not operate in cnidarians at the oral-aboral axis (Chiori et al., 2009) and that genetic changes over ~500 million years of evolution have obscured the relationship of Hox- and ParaHox-related gene function in this phylum to that in bilaterians (Chourrout et al., 2006; Finnerty et al., 2004; Kamm et al., 2006; Thomas-Chollier et al., 2010).
Whereas studies of Hox genes have not been as illuminating as originally hoped with regard to the evolution of axes in metazoans, studies of the Wnt signaling pathway have been. In Hydra, from which genes encoding most of the components of the canonical Wnt pathway have been cloned, seven of the ten Wnt genes identified in Hydra are expressed in the hypostome (the oral dome of the polyp) (Hobmayer et al., 2000; Lengfeld et al., 2009). Moreover, all of the Wnt genes investigated show a staggered spatial expression pattern along the oral-aboral axis of the Nematostella planula larva and the Hydra polyp, suggesting that they are involved in the patterning of this axis (Guder et al., 2006; Kusserow et al., 2005; Lee et al., 2006). However, whether the role of Wnt signaling is to pattern and specify the axis in a Hox-like manner or to control gastrulation and endoderm formation remains a matter of debate, as canonical Wnt pathway activation by LiCl also leads to an expansion of the endoderm during gastrulation (Wikramanayake et al., 2003).
In Clytia, two maternally expressed Frizzled Wnt receptors localize to opposing ends of the egg, where they act to define the oral-aboral axis (Momose and Houliston, 2007). In addition, a maternally expressed Wnt gene accounts, in part, for the observed role of canonical Wnt signaling during early embryonic axis formation in Clytia (Momose et al., 2008). Wnt signaling also appears to play an important role in axial patterning in the embryo and polyp of Hydractinia echinata (Duffy et al., 2010; Plickert et al., 2006). Recent evidence from Hydra suggests an interesting link between canonical and non-canonical Wnt signaling during bud formation (Philipp et al., 2009). In addition to Wnt receptors and ligands, several intracellular components of the canonical Wnt signaling pathway, including Dishevelled and β-catenin, function in cnidarian axis formation and gastrulation (Gee et al., 2010; Lee et al., 2007). Interestingly, chemical perturbation of canonical and non-canonical Wnt signaling suggests that a hierarchical relationship exists between these two pathways during budding of Hydra (Philipp et al., 2009). A recent functional study on Strabismus suggests a crucial role for the Wnt planar cell polarity (PCP) pathway during gastrulation of Nematostella. (Kumburegama et al., 2011).
Thus, in cnidarians, Wnt signaling appears to play a decisive role in establishing the animal/oral pole that subsequently develops into the hypostomal organizer of the polyp. This seems to be one of the oldest conserved developmental mechanisms in animal evolution because in vertebrates and other organisms, canonical Wnt signaling is involved in defining the blastopore (see Glossary, Box 1) or a derivative of it (e.g. the ‘organizer’ in vertebrates) (reviewed by Weaver and Kimelman, 2004). Since Wnt signaling in vertebrates is crucial for posterior development, it is tempting to homologize the A-P axis of vertebrates with the aboral-oral axis of the cnidarians. However, unlike in cnidarians, extensive morphogenetic movements of the tissue during vertebrate gastrulation change the axial position of the cells: while the closing blastopore becomes the posterior end, early involuting cells of the dorsal blastopore lip have a dorsoanterior fate. Hence, it appears that the oral-aboral axis of cnidarians more likely corresponds to the vegetal-animal axis of vertebrates.
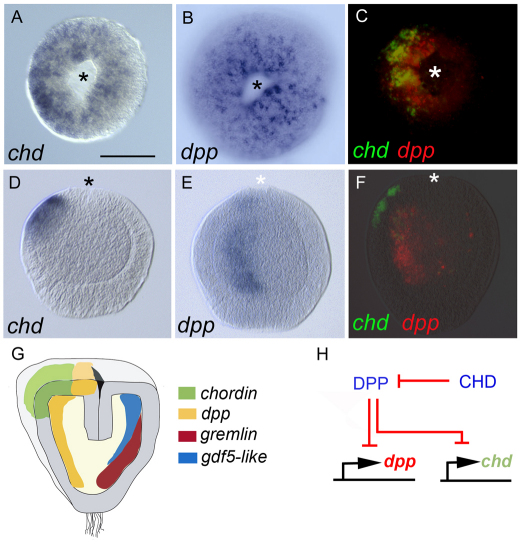
The dorsal-ventral (D-V) axis of bilaterians is established through the conserved functions of the signaling factor BMP2/4 (Dpp in Drosophila) together with the secreted BMP antagonist Chordin (Short gastrulation in Drosophila). Studying these genes in cnidarians has helped us understand the evolutionary history of the D-V axis. Components of the BMP pathway are expressed during embryogenesis in the anthozoans Nematostella and Acropora (Technau et al., 2005). Strikingly, in Acropora the bmp2/4 homolog is asymmetrically expressed (Hayward et al., 2002). Subsequently, it was found that bmp2/4, its co-factor bmp5-8, the BMP-like ligand gdf5, the antagonists chordin and gremlin1, as well as several other genes, such as most of the Hox genes, are expressed asymmetrically with respect to the oral-aboral axis in Nematostella (Fig. 6) (Finnerty et al., 2004; Hayward et al., 2002; Matus et al., 2006a; Matus et al., 2006b; Rentzsch et al., 2006). These findings demonstrate the existence of a molecularly defined second body axis, perpendicular to the oral-aboral body axis, called the directive axis. Surprisingly, in Nematostella bmp2/4 and chordin do not form opposing gradients of expression as they do in vertebrates or flies but instead are expressed on the same side after initially being expressed in a radial pattern around the blastopore (Rentzsch et al., 2006). Recent findings suggest that BMP and chordin function in a negative-feedback loop (Fig. 6H), indicating that BMP signaling is required for symmetry breaking to occur (Saina et al., 2009). It is at present unclear what the consequences of the molecular asymmetry of BMP signaling in anthozoans is, but during metamorphosis into the primary polyp, bmp2/4 expression becomes localized in all eight mesenteries, where it could regulate the differentiation of the retractor muscles (Finnerty et al., 2004; Saina and Technau, 2009). By contrast, in Hydra, in which no morphological asymmetry is detectable, chordin expression is radial in the adult polyp, whereas it is dynamically expressed during budding and regeneration. This suggests that the symmetry break caused by BMP signaling was either lost during evolution or reverted to a radial pattern in the polyp stage of Hydra, leading to a secondary radialization of the body plan (Rentzsch et al., 2007).
Fig. 6.
Symmetry break and asymmetric expression of BMP-like genes and BMP antagonists in Nematostella embryos. (A,B) Early gastrula stage (oral view) showing radial expression of a BMP antagonist, the Nematostella homolog chordin (chd, A), and of the Nematostella BMP2 homolog dpp (B). (C) Double in situ hybridization of chordin and dpp showing that during the mid-gastrula stage, a symmetry break occurs and both genes become expressed on the same side of the blastopore. (D,E) During the planula stage, expression of chordin remains lateral to the blastopore (D), whereas dpp is largely expressed in an endodermal stripe and in a spot at the border of the blastopore (E), on the side of chordin expression. (F) Double in situ hybridization of chordin and dpp in a planula larva showing that both genes remain expressed asymmetrically, on the same side, but segregate with respect to ectoderm and endoderm. Asterisks mark the blastopore. Scale bar: 100 μm. (G) Schematic of the planula stage illustrating the asymmetric expression of chordin and dpp on one side, and of gdf5-like, a member of the BMP family, and of gremlin, a BMP antagonist, on the opposite side. Note that a number of other genes (not shown for clarity) are also expressed asymmetrically, indicative of a directive axis. (H) Double negative-feedback loop between Dpp and Chordin as suggested by morpholino-mediated gene knockdown experiments (Saina et al., 2009). Images in A-G are reproduced with permission (Rentzsch et al., 2006).
Chordin is an important component of the Spemann-Mangold organizer (see Glossary, Box 1) and, therefore, the expression of chordin and Wnt genes around the cnidarian blastopore suggests that the cnidarian and bilaterian organizers are homologous. Accordingly, in division experiments with Nematostella embryos, only the oral half can regenerate a normal polyp (Fritzenwanker et al., 2007; Lee et al., 2007). Furthermore, transplantation of part of the Nematostella blastopore lip from an early gastrula to an aboral position induces the outgrowth of a second oral-aboral axis (Kraus et al., 2007), indicating that the cnidarian blastopore (or part of it) is homologous to the blastoporal organizer of vertebrates. As first reported in 1909, organizer activity is also present at the oral end of the Hydra polyp, at the hypostome, which directly develops from the embryonic blastopore (Browne, 1909). Wnt signaling is crucial for the organizing activity of the hypostome, as upregulation of the canonical Wnt pathway in Hydra results in ectopic head formation in the body column (Broun and Bode, 2002; Broun et al., 2005; Gee et al., 2010).
In summary, BMPs and chordin (or other BMP-binding molecules) are components of an ancient molecular system used to generate axial asymmetries. Since the morphological consequences of the deployment of this system in cnidarians are drastically different from those in bilaterians, it is premature to homologize the cnidarian directive axis with the D-V axis of bilaterians. Nonetheless, it is clear that the common ancestor of cnidarians and bilaterians used this signaling system for axial differentiation.
Can cnidarians inform us about the evolution of the mesoderm?
Cnidarians, being diploblasts, lack the third germ layer, the mesoderm. To trace the evolutionary origin of the mesoderm, researchers have searched for cnidarian homologs of genes involved in bilaterian mesoderm formation. Most of these genes encode transcription factors, such as the bHLH protein Twist, the zinc-finger protein Snail, the T-box factor Brachyury, myocyte enhancer factor 2 (Mef2) and the HMG protein Forkhead/FoxA, and virtually all of them are present in cnidarians and show differential expression during embryogenesis or later developmental processes. Interestingly, nearly all of these genes appear to be expressed at the blastopore (or the hypostome) and in all, or part, of the endoderm (Fritzenwanker et al., 2004; Hayward et al., 2004; Martindale et al., 2004; Matus et al., 2006b; Scholz and Technau, 2003; Spring et al., 2002; Spring et al., 2000; Technau and Bode, 1999; Technau and Scholz, 2003). This suggests that mesoderm might have arisen from endomesoderm (see Glossary, Box 1) in the common ancestor of bilaterians by an altered combination of interactions between these developmental regulators; in fact, Hydra Brachyury can induce mesoderm in Xenopus (Marcellini et al., 2003), suggesting that it is not the gene but rather the regulatory context that has evolved.
Since gastrulation is tightly linked to germ layer formation, researchers have also begun to investigate the molecular basis of gastrulation in cnidarians (Fritzenwanker et al., 2004; Kumburegama et al., 2011; Magie et al., 2007). Interestingly, virtually all possible modes of gastrulation (invagination, immigration, epiboly, delamination) occur in cnidarians (Tardent, 1978). With the advent of transgenic technology, it should be possible to follow individual labeled cells in a cnidarian embryo and to monitor their morphogenetic behavior during gastrulation in normal and experimentally manipulated embryos, so as to provide insights into the evolutionary basis of gastrulation movements and their molecular underpinnings.
Do the stem cells found in cnidarians share features with vertebrate stem cells?
Interest in stem cells has increased greatly recently owing to their therapeutic potential. However, because most studies have concentrated on vertebrate models we still have a lot to learn regarding the evolution of stem cells. Studies in cnidarians, particularly Hydra and other hydrozoans, are especially relevant to our understanding of stem cell evolution. Hydra has three cell lineages, which are all self-renewing and maintained by stem cells. The two epithelial cell lineages (ectodermal and endodermal) are maintained by division of cells in the body column. Thus, the differentiated epithelial cells of the body column also serve as stem cells. The interstitial cell lineage of Hydra consists of a multipotent stem cell population that gives rise to nerves, secretory cells, nematocytes and germ cells. Initial attempts to determine the evolutionary relationship between cnidarian and vertebrate stem cells involved searching sequenced cnidarian genomes and EST datasets for homologs of the four pluripotency genes that are known to be expressed in vertebrate stem cells (Klf4, Oct4, Sox2 and Nanog). Clear homologs of these genes have not been identified in the Nematostella or Hydra genome (Chapman et al., 2010), suggesting that either the role of these key genes is performed by related genes or, alternatively, that the circuitry for producing stem cells evolved independently in cnidarians and vertebrates. Support for the latter scenario comes from observations suggesting that the interstitial cell lineage is only present in hydrozoans. Identification of the genes that maintain ‘stemness’ in Hydra and other hydrozoans is an important goal for understanding whether cnidarian and vertebrate stem cells share any evolutionary history.
Limitations and future directions
For many years, studies of cnidarian development, particularly of pattern formation, stem cells and regeneration, were dominated by research using the adult Hydra polyp, allowing only indirect comparisons with bilaterian embryonic development. However, molecular studies of cnidarian embryos and larvae have gained momentum with the introduction of Nematostella and Clytia as models. We expect that the recent technical advances in these systems will fuel research to better understand axis and germ layer evolution and to understand the origin of stem cells and neurogenesis. We also expect that, as more cnidarians are developed as models, particularly from taxonomic groups that have been little studied to date, such as scyphozoans, cubozoans and scleractinian corals, we will begin to understand the molecular basis for the dramatic morphological variation that exists among cnidarian lineages. Comparisons between these morphologically diverse species might provide insights into the constraints of the underlying developmental programs. Even though most cnidarians will never become laboratory models, their genomes hold important information regarding the evolution of developmental pathways in bilaterians. Thus, a goal for the future is the generation of draft genome sequences for more cnidarian species, followed by comparative analyses to identify conserved and diverged features of their gene sets.
Although the recent development of methods for genetically manipulating cnidarians has facilitated studies of gene function, many of the tools available for more mature model organisms are still lacking for cnidarians. A more robust RNAi approach and improved transgenic methods (e.g. with inducible promoters or landing sites for recombination) are needed. The identification of more cell- or tissue-specific promoters to drive the expression of fluorescent protein genes will enable morphogenetic processes, such as gastrulation and nervous system restructuring during regeneration, to be followed in vivo by four-dimensional confocal microscopy.
It remains to be seen whether classical genetic screens can be performed in cnidarians. Early attempts at this in Hydra were hampered by the low numbers of embryos that can be obtained and by the lengthy embryonic dormancy that most Hydra strains undergo. None of the responsible genes has been identified for the 39 existing Hydra mutants (Sugiyama and Fujisawa, 1978). Ongoing inbreeding programs with defined Nematostella strains might make mutant screens feasible in the future, but Nematostella has a relatively long generation time of 4-6 months. In this respect, the hydrozoan Clytia, which has a generation time of 3-4 weeks, perhaps holds more promise. Genetic mapping and cloning of the histocompatibility complex has been carried out in the hydrozoan Hydractinia (Nicotra et al., 2009), indicating that genetic approaches are feasible in cnidarians.
Although studies of cnidarian development to date have focused on axis formation and regeneration, cnidarian models offer exciting opportunities for investigating other aspects of development. For example, how development of the cnidarian nervous system is controlled and how its development relates to nervous system development in bilaterians are of obvious interest. Future efforts will certainly expand on recent studies using cnidarians to understand eye evolution, as cnidarians are the only animals among the four basal metazoan phyla that have evolved sophisticated eyes. Whereas many hydrozoan medusa have simple eyes at the rim of the bell, the most sophisticated eyes are found in the cubozoan jellyfish. Box jellyfish have a total of 24 eyes arranged in a set of four rhopalia, each consisting of two lens eyes and four bilaterally paired pigment cup eyes (Kozmik et al., 2008; O'Connor et al., 2009). Cnidarians have multiple ciliary opsins, the light-sensitive receptors found in photoreceptor cells (Suga et al., 2008). Furthermore, although they do not possess a bona-fide Pax6 gene, which is the so-called master regulator of eye development in bilaterians (Gehring and Ikeo, 1999), they have a related, putative ancestral PaxB gene that is likely to function in eye development (Kozmik et al., 2003; Nilsson, 2009; Nilsson et al., 2005; Suga et al., 2010). As the molecular circuitry underlying eye formation in cnidarians is defined in more detail, the degree to which eye development in cnidarians and bilaterians is evolutionarily related should finally be revealed. Since even eyeless cnidarians respond to light, the identification of the light-sensitive cells in such cnidarians should yield insights into the evolutionary origins of light-sensing organs in general.
In summary, with the availability of genome sequences and the advent of gene knockdown techniques and transgenics it is now possible to carry out experimental studies of developmental processes in cnidarians that would have been impossible only a few years ago. We look forward to finding the answers to long-standing questions, gaining new insights and revealing surprising findings from the continued study of these remarkable animals.
Acknowledgements
We thank David Miller, Zoe Richards and Eldon Ball for photographs of Acropora; Thomas Holstein for a picture of a Hydra polyp; Hanna Kraus for photographs of Clytia hemisphaerica; Hanna Kraus for the drawings of the Nematostella, Clytia and Acropora life cycles and the Hydra polyp in Fig. 2; Yulia Kraus for help with Fig. 4; Catherine Dana, Thomas Bosch and Kostya Khalthurin for photographs of transgenic Hydra; Günter Plickert for the transgenic Hydractinia images; and three anonymous reviewers for constructive comments. We apologize to all cnidarian researchers whose work could not be cited owing to space constraints. Research in the U.T. lab is supported by the Austrian Science foundation FWF, the Norwegian Research Council NFR and the EU-Marie-Curie Initial training network EVONET. Research in the R.E.S. lab is supported by the NIH National Institute of General Medical Sciences. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- Ball E. E., Hayward D. C., Reece-Hoyes J. S., Hislop N. R., Samuel G., Saint R., Harrison P. L., Miller D. J. (2002). Coral development: from classical embryology to molecular control. Int. J. Dev. Biol. 46, 671-678 [PubMed] [Google Scholar]
- Ball E. E., Hayward D. C., Saint R., Miller D. J. (2004). A simple plan-cnidarians and the origins of developmental mechanisms. Nat. Rev. Genet. 5, 567-577 [DOI] [PubMed] [Google Scholar]
- Bielen H., Oberleitner S., Marcellini S., Gee L., Lemaire P., Bode H. R., Rupp R., Technau U. (2007). Divergent functions of two ancient Hydra Brachyury paralogues suggest specific roles for their C-terminal domains in tissue fate induction. Development 134, 4187-4197 [DOI] [PubMed] [Google Scholar]
- Bode H. R. (1992). Continuous conversion of neuron phenotype in hydra. Trends Genet. 8, 279-284 [DOI] [PubMed] [Google Scholar]
- Bode H. R. (1996). The interstitial cell lineage of hydra: a stem cell system that arose early in evolution. J. Cell Sci. 109, 1155-1164 [DOI] [PubMed] [Google Scholar]
- Bode P. M., Bode H. R. (1980). Formation of pattern in regenerating tissue pieces of hydra attenuata. I. Head-body proportion regulation. Dev. Biol. 78, 484-496 [DOI] [PubMed] [Google Scholar]
- Bosch T. C. (2009). Hydra and the evolution of stem cells. BioEssays 31, 478-486 [DOI] [PubMed] [Google Scholar]
- Bottger A., Alexandrova O., Cikala M., Schade M., Herold M., David C. N. (2002). GFP expression in Hydra: lessons from the particle gun. Dev. Genes Evol. 212, 302-305 [DOI] [PubMed] [Google Scholar]
- Bottger A., Strasser D., Alexandrova O., Levin A., Fischer S., Lasi M., Rudd S., David C. N. (2006). Genetic screen for signal peptides in Hydra reveals novel secreted proteins and evidence for non-classical protein secretion. Eur. J. Cell Biol. 85, 1107-1117 [DOI] [PubMed] [Google Scholar]
- Brauer A. (1891). Über die Entwicklung von Hydra. Z. Wiss. Zool. 52, 169-216 [Google Scholar]
- Broun M., Bode H. R. (2002). Characterization of the head organizer in hydra. Development 129, 875-884 [DOI] [PubMed] [Google Scholar]
- Broun M., Gee L., Reinhardt B., Bode H. R. (2005). Formation of the head organizer in hydra involves the canonical Wnt pathway. Development 132, 2907-2916 [DOI] [PubMed] [Google Scholar]
- Browne E. (1909). The production of new hydrants by the insertion of small grafts. J. Exp. Zool. 7, 1-37 [Google Scholar]
- Burton P. M., Finnerty J. R. (2009). Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis. Dev. Genes Evol. 219, 79-87 [DOI] [PubMed] [Google Scholar]
- Campbell R. D. (1976). Elimination by Hydra interstitial and nerve cells by means of colchicine. J. Cell Sci. 21, 1-13 [DOI] [PubMed] [Google Scholar]
- Campbell R. D., David C. N. (1974). Cell cycle kinetics and development of Hydra attenuata. II. Interstitial cells. J. Cell Sci. 16, 349-358 [DOI] [PubMed] [Google Scholar]
- Cartwright P., Halgedahl S. L., Hendricks J. R., Jarrard R. D., Marques A. C., Collins A. G., Lieberman B. S. (2007). Exceptionally preserved jellyfishes from the Middle Cambrian. PLoS One 2, e1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. A., Kirkness E. F., Simakov O., Hampson S. E., Mitros T., Weinmaier T., Rattei T., Balasubramanian P. G., Borman J., Busam D., et al. (2010). The dynamic genome of Hydra. Nature 464, 592-596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera S., Ghila L., Dobretz K., Wenger Y., Bauer C., Buzgariu W., Martinou J. C., Galliot B. (2009). Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev. Cell 17, 279-289 [DOI] [PubMed] [Google Scholar]
- Chiori R., Jager M., Denker E., Wincker P., Da Silva C., Le Guyader H., Manuel M., Queinnec E. (2009). Are Hox genes ancestrally involved in axial patterning? Evidence from the hydrozoan Clytia hemisphaerica (Cnidaria). PLoS One 4, e4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourrout D., Delsuc F., Chourrout P., Edvardsen R. B., Rentzsch F., Renfer E., Jensen M. F., Zhu B., de Jong P., Steele R. E., et al. (2006). Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature 442, 684-687 [DOI] [PubMed] [Google Scholar]
- Collins A. G. (2002). Phylogeny of Medusozoa and the evolution of cnidarian life cycles. J. Evol. Biol. 15, 418-432 [Google Scholar]
- Collins A. G., Bentlage B., Matsumoto G. I., Haddock S. H., Osborn K. J., Schierwater B. (2006). Solution to the phylogenetic enigma of Tetraplatia, a worm-shaped cnidarian. Biol. Lett. 2, 120-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte D., Jr, Mello C. C. (2003). RNA interference in Caenorhabditis elegans. In Current Protocols in Molecular Biology, Chapter 26, Unit 26.3 [DOI] [PubMed] [Google Scholar]
- Cummings S. G., Bode H. R. (1984). Head regeneration and polarity reversal in Hydra attenuata can occur in the absence of DNA synthesis. Rouxs Arch. Dev. Biol. 194, 79-86 [DOI] [PubMed] [Google Scholar]
- Darling J. A., Reitzel A. M., Finnerty J. R. (2004). Regional population structure of a widely introduced estuarine invertebrate: Nematostella vectensis Stephenson in New England. Mol. Ecol. 13, 2969-2981 [DOI] [PubMed] [Google Scholar]
- Darling J. A., Reitzel A. R., Burton P. M., Mazza M. E., Ryan J. F., Sullivan J. C., Finnerty J. R. (2005). Rising starlet: the starlet sea anemone, Nematostella vectensis. BioEssays 27, 211-221 [DOI] [PubMed] [Google Scholar]
- David C. N., Campbell R. D. (1972). Cell cycle kinetics and development of Hydra attenuata. I. Epithelial cells. J. Cell Sci. 11, 557-568 [DOI] [PubMed] [Google Scholar]
- David C. N., Gierer A. (1974). Cell cycle kinetics and development of Hydra attenuata. III. Nerve and nematocyte differentiation. J. Cell Sci. 16, 359-375 [DOI] [PubMed] [Google Scholar]
- David C. N., Murphy S. (1977). Characterization of interstitial stem cells in hydra by cloning. Dev. Biol. 58, 372-383 [DOI] [PubMed] [Google Scholar]
- Denker E., Manuel M., Leclere L., Le Guyader H., Rabet N. (2008). Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa, Cnidaria). Dev. Biol. 315, 99-113 [DOI] [PubMed] [Google Scholar]
- Duffy D. J., Plickert G., Kuenzel T., Tilmann W., Frank U. (2010). Wnt signaling promotes oral but suppresses aboral structures in Hydractinia metamorphosis and regeneration. Development 137, 3057-3066 [DOI] [PubMed] [Google Scholar]
- Dunn S. R., Phillips W. S., Green D. R., Weis V. M. (2007). Knockdown of actin and caspase gene expression by RNA interference in the symbiotic anemone Aiptasia pallida. Biol. Bull 212, 250-258 [DOI] [PubMed] [Google Scholar]
- Faurot L. (1895). L'anatomie, l'histologie, et le développement des Actinies. Arch. Zool. Exp. Gén. Sér. 3 3, 43-262 [Google Scholar]
- Faurot L. (1903). Développement du pharynx des couples et des paires de cloisons chez les Hexactinies. Arch. Zool. Exp. Gén. Sér. 3 1, 359-399 [Google Scholar]
- Finnerty J. R., Pang K., Burton P., Paulson D., Martindale M. Q. (2004). Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science 304, 1335-1337 [DOI] [PubMed] [Google Scholar]
- Foret S., Knack B., Houliston E., Momose T., Manuel M., Queinnec E., Hayward D. C., Ball E. E., Miller D. J. (2010). New tricks with old genes: the genetic bases of novel cnidarian traits. Trends Genet. 26, 154-158 [DOI] [PubMed] [Google Scholar]
- Freeman G. (1990). The establishment and role of polarity during embryogenesis in hydrozoans. In The Cellular and Molecular Biology of Pattern Formation (ed. Stocum D. L., Karr T. L.), pp. 3-30 Oxford: Oxford University Press; [Google Scholar]
- Fritzenwanker J. H., Technau U. (2002). Induction of gametogenesis in the basal cnidarian Nematostella vectensis(Anthozoa). Dev. Genes Evol. 212, 99-103 [DOI] [PubMed] [Google Scholar]
- Fritzenwanker J. H., Saina M., Technau U. (2004). Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev. Biol. 275, 389-402 [DOI] [PubMed] [Google Scholar]
- Fritzenwanker J. H., Genikhovich G., Kraus Y., Technau U. (2007). Early development and axis specification in the sea anemone Nematostella vectensis. Dev. Biol. 310, 264-279 [DOI] [PubMed] [Google Scholar]
- Galliot B. (2000). Conserved and divergent genes in apex and axis development of cnidarians. Curr. Opin. Genet. Dev. 10, 629-637 [DOI] [PubMed] [Google Scholar]
- Galliot B., Miller D. (2000). Origin of anterior patterning. How old is our head? Trends Genet. 16, 1-5 [DOI] [PubMed] [Google Scholar]
- Galliot B., Tanaka E., Simon A. (2008). Regeneration and tissue repair: themes and variations. Cell. Mol. Life Sci. 65, 3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B., Quiquand M., Ghila L., de Rosa R., Miljkovic-Licina M., Chera S. (2009). Origins of neurogenesis, a cnidarian view. Dev. Biol. 332, 2-24 [DOI] [PubMed] [Google Scholar]
- Gee L., Hartig J., Law L., Wittlieb J., Khalturin K., Bosch T. C., Bode H. R. (2010). beta-catenin plays a central role in setting up the head organizer in hydra. Dev. Biol. 340, 116-124 [DOI] [PubMed] [Google Scholar]
- Gehring W. J., Ikeo K. (1999). Pax6, mastering eye morphogenesis and eye evolution. Trends Genet. 15, 371-377 [DOI] [PubMed] [Google Scholar]
- Genikhovich G., Technau U. (2009). The starlet sea anemone Nematostella vectensis: an anthozoan model organism for studies in comparative genomics and functional evolutionary developmental biology. Cold Spring Harb. Protoc. 2009, pdb emo129 [DOI] [PubMed] [Google Scholar]
- Gierer A., Meinhardt H. (1972). A theory of biological pattern formation. Kybernetik 12, 30-39 [DOI] [PubMed] [Google Scholar]
- Gierer A., Berking S., Bode H., David C. N., Flick K., Hansmann G., Schaller H., Trenkner E. (1972). Regeneration of hydra from reaggregated cells. Nat. New Biol. 239, 98-101 [DOI] [PubMed] [Google Scholar]
- Guder C., Philipp I., Lengfeld T., Watanabe H., Hobmayer B., Holstein T. W. (2006). The Wnt code: cnidarians signal the way. Oncogene 25, 7450-7460 [DOI] [PubMed] [Google Scholar]
- Hand C., Uhlinger K. R. (1992). The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biol. Bull. 182, 169-176 [DOI] [PubMed] [Google Scholar]
- Hassel M., Bieller A. (1996). Stepwise transfer from high to low lithium concentrations increases the head-forming potential in Hydra vulgaris and possibly activates the PI cycle. Dev. Biol. 177, 439-448 [DOI] [PubMed] [Google Scholar]
- Hayward D. C., Samuel G., Pontynen P. C., Catmull J., Saint R., Miller D. J., Ball E. E. (2002). Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc. Natl. Acad. Sci. USA 99, 8106-8111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D. C., Miller D. J., Ball E. E. (2004). snail expression during embryonic development of the coral Acropora: blurring the diploblast/triploblast divide? Dev. Genes Evol. 214, 257-260 [DOI] [PubMed] [Google Scholar]
- Heimfeld S., Bode H. R. (1984). Interstitial cell migration in Hydra attenuata. I. Quantitative description of cell movements. Dev. Biol. 105, 1-9 [DOI] [PubMed] [Google Scholar]
- Hobmayer B., Rentzsch F., Kuhn K., Happel C. M., von Laue C. C., Snyder P., Rothbacher U., Holstein T. W. (2000). WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407, 186-189 [DOI] [PubMed] [Google Scholar]
- Holstein T. (1981). The morphogenesis of nematocytes in Hydra and Forskalia: an ultrastructural study. J. Ultrastruct. Res. 75, 276-290 [DOI] [PubMed] [Google Scholar]
- Holstein T. W., Hobmayer E., David C. N. (1991). Pattern of epithelial cell cycling in hydra. Dev. Biol. 148, 602-611 [DOI] [PubMed] [Google Scholar]
- Holstein T. W., Hobmayer E., Technau U. (2003). Cnidarians: an evolutionarily conserved model system for regeneration? Dev. Dyn. 226, 257-267 [DOI] [PubMed] [Google Scholar]
- Houliston E., Momose T., Manuel M. (2010). Clytia hemisphaerica: a jellyfish cousin joins the laboratory. Trends Genet. 26, 159-167 [DOI] [PubMed] [Google Scholar]
- Hwang J. S., Takaku Y., Momose T., Adamczyk P., Ozbek S., Ikeo K., Khalturin K., Hemmrich G., Bosch T. C., Holstein T. W., et al. (2010). Nematogalectin, a nematocyst protein with GlyXY and galectin domains, demonstrates nematocyte-specific alternative splicing in Hydra. Proc. Natl. Acad. Sci. USA 107, 18539-18544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L. H. (1940). The Invertebrates: Protozoa through Ctenophora. New York: McGraw-Hill; [Google Scholar]
- Jägersten G. (1955). On the early phylogeny of the Bilateria. Zool. Bidrag. Uppsala 30, 321-354 [Google Scholar]
- Kamm K., Schierwater B., Jakob W., Dellaporta S. L., Miller D. J. (2006). Axial patterning and diversification in the cnidaria predate the Hox system. Curr. Biol. 16, 920-926 [DOI] [PubMed] [Google Scholar]
- Kasbauer T., Towb P., Alexandrova O., David C. N., Dall'armi E., Staudigl A., Stiening B., Bottger A. (2007). The Notch signaling pathway in the cnidarian Hydra. Dev. Biol. 303, 376-390 [DOI] [PubMed] [Google Scholar]
- Khalturin K., Anton-Erxleben F., Milde S., Plotz C., Wittlieb J., Hemmrich G., Bosch T. C. (2007). Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev. Biol. 309, 32-44 [DOI] [PubMed] [Google Scholar]
- Khalturin K., Anton-Erxleben F., Sassmann S., Wittlieb J., Hemmrich G., Bosch T. C. (2008). A novel gene family controls species-specific morphological traits in Hydra. PLoS Biol. 6, e278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K., Hemmrich G., Fraune S., Augustin R., Bosch T. C. (2009). More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 25, 404-413 [DOI] [PubMed] [Google Scholar]
- Kortschak R. D., Samuel G., Saint R., Miller D. J. (2003). EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr. Biol. 13, 2190-2195 [DOI] [PubMed] [Google Scholar]
- Kozmik Z., Daube M., Frei E., Norman B., Kos L., Dishaw L. J., Noll M., Piatigorsky J. (2003). Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev. Cell 5, 773-785 [DOI] [PubMed] [Google Scholar]
- Kozmik Z., Ruzickova J., Jonasova K., Matsumoto Y., Vopalensky P., Kozmikova I., Strnad H., Kawamura S., Piatigorsky J., Paces V., et al. (2008). Assembly of the cnidarian camera-type eye from vertebrate-like components. Proc. Natl. Acad. Sci. USA 105, 8989-8993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus Y., Fritzenwanker J. H., Genikhovich G., Technau U. (2007). The blastoporal organiser of a sea anemone. Curr. Biol. 17, R874-R876 [DOI] [PubMed] [Google Scholar]
- Kumburegama S., Wijesena N., Xu R., Wikramanayake A. H. (2011). Strabismus-mediated primary archenteron invagination is uncoupled from Wnt/SZ-catenin-dependent endoderm cell fate specification in Nematostella vectensis (Anthozoa, Cnidaria): implications for the evolution of gastrulation. Evodevo 2, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzel T., Heiermann R., Frank U., Muller W., Tilmann W., Bause M., Nonn A., Helling M., Schwarz R. S., Plickert G. (2010). Migration and differentiation potential of stem cells in the cnidarian Hydractinia analysed in eGFP-transgenic animals and chimeras. Dev. Biol. 348, 120-129 [DOI] [PubMed] [Google Scholar]
- Kusserow A., Pang K., Sturm C., Hrouda M., Lentfer J., Schmidt H. A., Technau U., von Haeseler A., Hobmayer B., Martindale M. Q., et al. (2005). Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156-160 [DOI] [PubMed] [Google Scholar]
- Lee P. N., Pang K., Matus D. Q., Martindale M. Q. (2006). A WNT of things to come: evolution of Wnt signaling and polarity in cnidarians. Semin. Cell Dev. Biol. 17, 157-167 [DOI] [PubMed] [Google Scholar]
- Lee P. N., Kumburegama S., Marlow H. Q., Martindale M. Q., Wikramanayake A. H. (2007). Asymmetric developmental potential along the animal-vegetal axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by Dishevelled. Dev. Biol. 310, 169-186 [DOI] [PubMed] [Google Scholar]
- Lengfeld T., Watanabe H., Simakov O., Lindgens D., Gee L., Law L., Schmidt H. A., Ozbek S., Bode H., Holstein T. W. (2009). Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Biol. 330, 186-199 [DOI] [PubMed] [Google Scholar]
- Lohmann J. U., Endl I., Bosch T. C. (1999). Silencing of developmental genes in Hydra. Dev. Biol. 214, 211-214 [DOI] [PubMed] [Google Scholar]
- MacWilliams H. K. (1983a). Hydra transplantation phenomena and the mechanism of hydra head regeneration. I. Properties of the head inhibition. Dev. Biol. 96, 217-238 [DOI] [PubMed] [Google Scholar]
- MacWilliams H. K. (1983b). Hydra transplantation phenomena and the mechanism of Hydra head regeneration. II. Properties of the head activation. Dev. Biol. 96, 239-257 [DOI] [PubMed] [Google Scholar]
- Magie C. R., Daly M., Martindale M. Q. (2007). Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev. Biol. 305, 483-497 [DOI] [PubMed] [Google Scholar]
- Marcellini S., Technau U., Smith J. C., Lemaire P. (2003). Evolution of Brachyury proteins: identification of a novel regulatory domain conserved within Bilateria. Dev. Biol. 260, 352-361 [DOI] [PubMed] [Google Scholar]
- Martin V. J., Littlefield C. L., Archer W. E., Bode H. R. (1997). Embryogenesis in hydra. Biol. Bull. 192, 345-363 [DOI] [PubMed] [Google Scholar]
- Martindale M. Q., Pang K., Finnerty J. R. (2004). Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa). Development 131, 2463-2474 [DOI] [PubMed] [Google Scholar]
- Martinez D. E., Iniguez A. R., Percell K. M., Willner J. B., Signorovitch J., Campbell R. D. (2010). Phylogeny and biogeography of Hydra (Cnidaria: Hydridae) using mitochondrial and nuclear DNA sequences. Mol. Phylogenet. Evol. 57, 403-410 [DOI] [PubMed] [Google Scholar]
- Matus D. Q., Pang K., Marlow H., Dunn C. W., Thomsen G. H., Martindale M. Q. (2006a). Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc. Natl. Acad. Sci. USA 103, 11195-11200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus D. Q., Thomsen G. H., Martindale M. Q. (2006b). Dorso/ventral genes are asymmetrically expressed and involved in germ-layer demarcation during cnidarian gastrulation. Curr. Biol. 16, 499-505 [DOI] [PubMed] [Google Scholar]
- Miljkovic-Licina M., Chera S., Ghila L., Galliot B. (2007). Head regeneration in wild-type hydra requires de novo neurogenesis. Development 134, 1191-1201 [DOI] [PubMed] [Google Scholar]
- Miller D. J., Ball E. E. (2008). Cryptic complexity captured: the Nematostella genome reveals its secrets. Trends Genet. 24, 1-4 [DOI] [PubMed] [Google Scholar]
- Miller D. J., Ball E. E., Technau U. (2005). Cnidarians and ancestral genetic complexity in the animal kingdom. Trends Genet. 21, 536-539 [DOI] [PubMed] [Google Scholar]
- Miller M. A., Technau U., Smith K. M., Steele R. E. (2000). Oocyte development in Hydra involves selection from competent precursor cells. Dev. Biol. 224, 326-338 [DOI] [PubMed] [Google Scholar]
- Momose T., Schmid V. (2006). Animal pole determinants define oral-aboral axis polarity and endodermal cell-fate in hydrozoan jellyfish Podocoryne carnea. Dev. Biol. 292, 371-380 [DOI] [PubMed] [Google Scholar]
- Momose T., Houliston E. (2007). Two oppositely localised frizzled RNAs as axis determinants in a cnidarian embryo. PLoS Biol. 5, e70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose T., Derelle R., Houliston E. (2008). A maternally localised Wnt ligand required for axial patterning in the cnidarian Clytia hemisphaerica. Development 135, 2105-2113 [DOI] [PubMed] [Google Scholar]
- Muller W. A. (1990). Ectopic head and foot formation in Hydra: diacylglycerol-induced increase in positional value and assistance of the head in foot formation. Differentiation 42, 131-143 [DOI] [PubMed] [Google Scholar]
- Nicotra M. L., Powell A. E., Rosengarten R. D., Moreno M., Grimwood J., Lakkis F. G., Dellaporta S. L., Buss L. W. (2009). A hypervariable invertebrate allodeterminant. Curr. Biol. 19, 583-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D. E. (2009). The evolution of eyes and visually guided behaviour. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2833-2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D. E., Gislen L., Coates M. M., Skogh C., Garm A. (2005). Advanced optics in a jellyfish eye. Nature 435, 201-205 [DOI] [PubMed] [Google Scholar]
- O'Connor M., Garm A., Nilsson D. E. (2009). Structure and optics of the eyes of the box jellyfish Chiropsella bronzie. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 195, 557-569 [DOI] [PubMed] [Google Scholar]
- Pankow S., Bamberger C. (2007). The p53 tumor suppressor-like protein nvp63 mediates selective germ cell death in the sea anemone Nematostella vectensis. PLoS One 2, e782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. V., Rogers A. D., Sheader M. (2002). The genetic structure of the rare lagoonal sea anemone, Nematostella vectensis Stephenson (Cnidaria; Anthozoa) in the United Kingdom based on RAPD analysis. Mol. Ecol. 11, 2285-2293 [DOI] [PubMed] [Google Scholar]
- Philipp I., Aufschnaiter R., Ozbek S., Pontasch S., Jenewein M., Watanabe H., Rentzsch F., Holstein T. W., Hobmayer B. (2009). Wnt/beta-catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra. Proc. Natl. Acad. Sci. USA 106, 4290-4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plickert G., Kroiher M., Munck A. (1988). Cell proliferation and early differentiation during embryonic development and metamorphosis of Hydractinia echinata. Development 103, 795-803 [DOI] [PubMed] [Google Scholar]
- Plickert G., Jacoby V., Frank U., Muller W. A., Mokady O. (2006). Wnt signaling in hydroid development: formation of the primary body axis in embryogenesis and its subsequent patterning. Dev. Biol. 298, 368-378 [DOI] [PubMed] [Google Scholar]
- Putnam N. H., Srivastava M., Hellsten U., Dirks B., Chapman J., Salamov A., Terry A., Shapiro H., Lindquist E., Kapitonov V. V., et al. (2007). Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86-94 [DOI] [PubMed] [Google Scholar]
- Renfer E., Amon-Hassenzahl A., Steinmetz P. R., Technau U. (2010). A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc. Natl. Acad. Sci. USA 107, 104-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F., Anton R., Saina M., Hammerschmidt M., Holstein T. W., Technau U. (2006). Asymmetric expression of the BMP antagonists chordin and gremlin in the sea anemone Nematostella vectensis: implications for the evolution of axial patterning. Dev. Biol. 296, 375-387 [DOI] [PubMed] [Google Scholar]
- Rentzsch F., Guder C., Vocke D., Hobmayer B., Holstein T. W. (2007). An ancient chordin-like gene in organizer formation of Hydra. Proc. Natl. Acad. Sci. USA 104, 3249-3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F., Fritzenwanker J. H., Scholz C. B., Technau U. (2008). FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development 135, 1761-1769 [DOI] [PubMed] [Google Scholar]
- Saina M., Technau U. (2009). Characterization of myostatin/gdf8/11 in the starlet sea anemone Nematostella vectensis. J. Exp. Zool. B Mol. Dev. Evol. 312, 780-788 [DOI] [PubMed] [Google Scholar]
- Saina M., Genikhovich G., Renfer E., Technau U. (2009). BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc. Natl. Acad. Sci. USA 106, 18592-18597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvini-Plawen L. V. (1978). On the origin and evolution of the lower Metazoa. Z. Zool. Syst. Evolut. Forsch. 16, 40-88 [Google Scholar]
- Schmid I., Tardent P. (1984). Migration of i-cells from ectoderm to endoderm in Hydra attenuata Pall. (Cnidaria, Hydrozoa) and their subsequent differentiation. Dev. Biol. 106, 469-477 [DOI] [PubMed] [Google Scholar]
- Scholz C. B., Technau U. (2003). The ancestral role of Brachyury: expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa). Dev. Genes Evol. 212, 563-570 [DOI] [PubMed] [Google Scholar]
- Sheader M., Suwailem A. M., Rowe G. A. (1997). The anemone, Nematostella vectensis, in Britain: considerations for conservation management. Aquat. Conserv. Mar. Freshw. Ecosyst. 7, 13-25 [Google Scholar]
- Siebert S., Thomsen S., Reimer M. M., Bosch T. C. (2005). Control of foot differentiation in Hydra: phylogenetic footprinting indicates interaction of head, bud and foot patterning systems. Mech. Dev. 122, 998-1007 [DOI] [PubMed] [Google Scholar]
- Smith K. M., Gee L., Bode H. R. (2000). HyAlx, an aristaless-related gene, is involved in tentacle formation in hydra. Development 127, 4743-4752 [DOI] [PubMed] [Google Scholar]
- Spring J., Yanze N., Middel A. M., Stierwald M., Groger H., Schmid V. (2000). The mesoderm specification factor twist in the life cycle of jellyfish. Dev. Biol. 228, 363-375 [DOI] [PubMed] [Google Scholar]
- Spring J., Yanze N., Josch C., Middel A. M., Winninger B., Schmid V. (2002). Conservation of Brachyury, Mef2, and Snail in the myogenic lineage of jellyfish: a connection to the mesoderm of bilateria. Dev. Biol. 244, 372-384 [DOI] [PubMed] [Google Scholar]
- Steele R. E., Dana C. E. (2009). Evolutionary history of the HAP2/GCS1 gene and sexual reproduction in metazoans. PLoS One 4, e7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele R. E., David C. N., Technau U. (2011). A genomic view of 500 million years of cnidarian evolution. Trends Genet. 27, 7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H., Schmid V., Gehring W. J. (2008). Evolution and functional diversity of jellyfish opsins. Curr. Biol. 18, 51-55 [DOI] [PubMed] [Google Scholar]
- Suga H., Tschopp P., Graziussi D. F., Stierwald M., Schmid V., Gehring W. J. (2010). Flexibly deployed Pax genes in eye development at the early evolution of animals demonstrated by studies on a hydrozoan jellyfish. Proc. Natl. Acad. Sci. USA 107, 14263-14268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Fujisawa T. (1978). Genetic analysis of developmental mechanisms in Hydra. II. Isolation and characterization of an interstitial cell-deficient strain. J. Cell Sci. 29, 35-52 [DOI] [PubMed] [Google Scholar]
- Sunagawa S., Wilson E. C., Thaler M., Smith M. L., Caruso C., Pringle J. R., Weis V. M., Medina M., Schwarz J. A. (2009). Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10, 258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Sugiyama T. (1984). Genetic analysis of developmental mechanisms in hydra. XII. Analysis of chimaeric hydra produced from a normal and a slow-budding strain (L4). J. Embryol. Exp. Morphol. 80, 155-173 [PubMed] [Google Scholar]
- Tardent P. (1978). Coelenterata, Cnidaria. Jena: Gustav Fischer Verlag; [Google Scholar]
- Tardent P., Holstein T. (1982). Morphology and morphodynamics of the stenotele nematocyst of Hydra attenuata Pall. (Hydrozoa, Cnidaria). Cell Tissue Res. 224, 269-290 [DOI] [PubMed] [Google Scholar]
- Technau U., Holstein T. W. (1992). Cell sorting during the regeneration of Hydra from reaggregated cells. Dev. Biol. 151, 117-127 [DOI] [PubMed] [Google Scholar]
- Technau U., Holstein T. W. (1996). Phenotypic maturation of neurons and continuous precursor migration in the formation of the peduncle nerve net in Hydra. Dev. Biol. 177, 599-615 [DOI] [PubMed] [Google Scholar]
- Technau U., Bode H. R. (1999). HyBra1, a Brachyury homologue, acts during head formation in Hydra. Development 126, 999-1010 [DOI] [PubMed] [Google Scholar]
- Technau U., Scholz C. B. (2003). Origin and evolution of endoderm and mesoderm. Int. J. Dev. Biol. 47, 531-539 [PubMed] [Google Scholar]
- Technau U., Cramer von Laue C., Rentzsch F., Luft S., Hobmayer B., Bode H. R., Holstein T. W. (2000). Parameters of self-organization in Hydra aggregates. Proc. Natl. Acad. Sci. USA 97, 12127-12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U., Miller M. A., Bridge D., Steele R. E. (2003). Arrested apoptosis of nurse cells during Hydra oogenesis and embryogenesis. Dev. Biol. 260, 191-206 [DOI] [PubMed] [Google Scholar]
- Technau U., Rudd S., Maxwell P., Gordon P. M., Saina M., Grasso L. C., Hayward D. C., Sensen C. W., Saint R., Holstein T. W., et al. (2005). Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 21, 633-639 [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M., Ledent V., Leyns L., Vervoort M. (2010). A non-tree-based comprehensive study of metazoan Hox and ParaHox genes prompts new insights into their origin and evolution. BMC Evol. Biol. 10, 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley A. (1744). Mémoires pour servir à l'histoire d'un genre de polypes d'eau douce. Leide: Jean and Herman Verbeek; [Google Scholar]
- Weaver C., Kimelman D. (2004). Move it or lose it: axis specification in Xenopus. Development 131, 3491-3499 [DOI] [PubMed] [Google Scholar]
- Wikramanayake A. H., Hong M., Lee P. N., Pang K., Byrum C. A., Bince J. M., Xu R., Martindale M. Q. (2003). An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature 426, 446-450 [DOI] [PubMed] [Google Scholar]
- Wittlieb J., Khalturin K., Lohmann J. U., Anton-Erxleben F., Bosch T. C. (2006). Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. USA 103, 6208-6211 [DOI] [PMC free article] [PubMed] [Google Scholar]