Fig. 2.
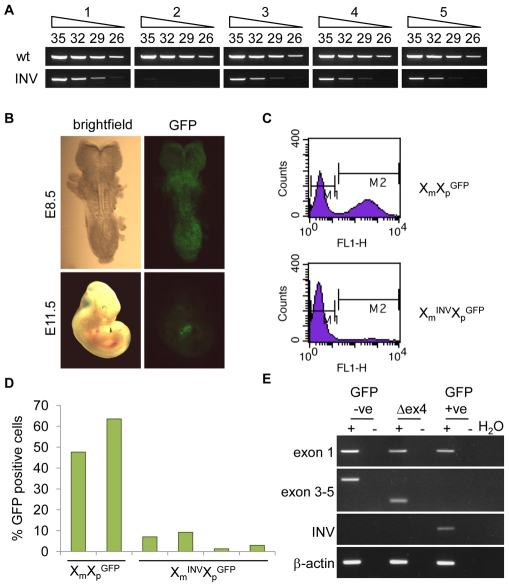
Heterozygous female embryos with a maternally transmitted XistINV allele exhibit secondary non-random X inactivation. (A) Allele-specific RT-PCR analysis of Xist from X INVmXp adult female kidneys (females 1-5). Top panel (wt) shows amplification with wild-type Xist allele specific primers (e and l) and the bottom panel presents amplification with XistINV specific primers (b and e). Numbers above the top panel show the number of cycles used. Note the six to nine cycle difference in amplification of wild-type versus XistINV fragments between different females. (B) Light and GFP microscopy images of E8.5 and E11.5 embryos with a maternally inherited XistINV allele and a paternally inherited GFP transgene. The diminishing proportion of green cells indicates secondary non-random X inactivation. (C) Examples of FACS histograms showing relative proportion of GFP-negative (M1) and GFP-positive (M2) cells in XmXp GFP and Xm INVXp GFP primary fibroblast cell lines established from E14.5 females. The x-axis represents the logarithmic level of GFP fluorescence and the y-axis shows the number of cells. 50,000 total events were acquired for each sample. (D) Quantitative analysis of FACS data obtained for two primary control fibroblast cultures (XmX GFPp) and for four primary fibroblast cultures from mutant heterozygous E14.5 embryos (Xm INVXp GFP). (E) RT-PCR analysis of the Xist transcripts produced by fibroblast cell lines expressing wild-type Xist, XistΔex4 or XistINV. Top panel, primers amplifying a sequence upstream of the inversion in exon 1 (a and h); second panel, primers situated in exon 3 and exon 5, respectively (e and m); third panel, primers flanking the inversion break-point that only amplify from the XistINV allele (b and e); bottom panel, β-actin loading control. + and − indicate reactions set up with and without reverse transcriptase, respectively. Positions of all Xist primers used are shown in Fig. S1 in the supplementary material.