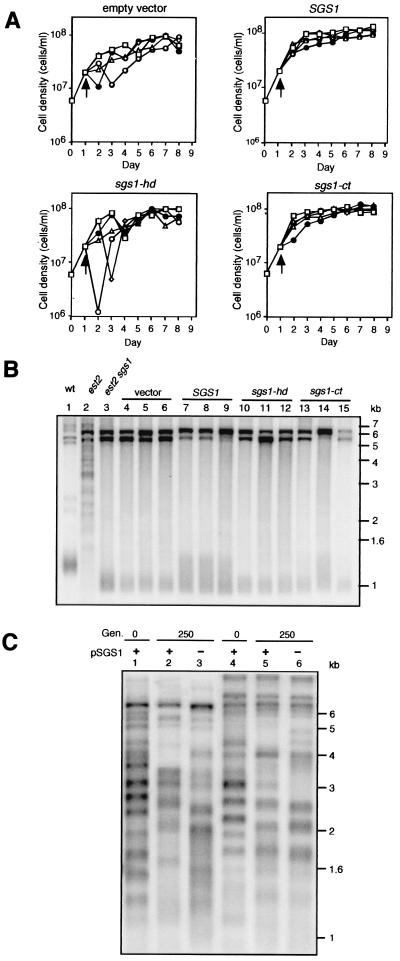
Figure 5.
Reintroduction of SGS1 into est2 sgs1 survivors restores growth rate and lengthens C1–3A/TG1–3 repeat tracts. The sgs1-hd (helicase-defective) allele carries a single amino acid substitution in the ATP binding domain (3). The sgs1-ct (Δ200) allele is a carboxyl-terminal truncation that removes the conserved RecQ carboxyl-terminal (ct) and RNaseD homology (HRD) domains (3). (A) Low copy plasmids carrying the SGS1, sgs1-hd, or sgs1-ct allele, or no insert, were introduced by transformation into est2 sgs1 survivors, 1 day after they were generated (indicated by arrow). Five random transformants were resuspended in medium lacking leucine and serially passaged each day between 1 × 105 cells/ml and 1.4 × 108 cells/ml. Each passage represented 10 generations of the wild-type strain. (B) Telomere structure analysis of strains in A on day 8. XhoI-digested genomic DNA was probed with a poly[AC/TG] sequence. Identical terminal band sizes were detected by probing blots for Y′ sequences (not shown). Lane 1, wild type; lane 2, est2 survivor; lane 3, est2 sgs1 survivor; lanes 4–6, vector only; lanes 7–9, SGS1; lanes 10–12, sgs1-hd; and lanes 13–15, sgs1-ct. (C) Telomere structure analysis of two independently derived est2 sgs1 type II survivors after the loss of plasmid-borne SGS1 (pSGS1). Lanes 1–3, survivor 1; and lanes 4–6, survivor 2. Lanes 1 and 4, telomeres before loss of pSGS1; lanes 2 and 5, 250 generations with selection for pSGS1; and lanes 3 and 6, 250 generations after loss of pSGS1. Genomic DNA was probed with a poly[AC/TG] sequence as in B.