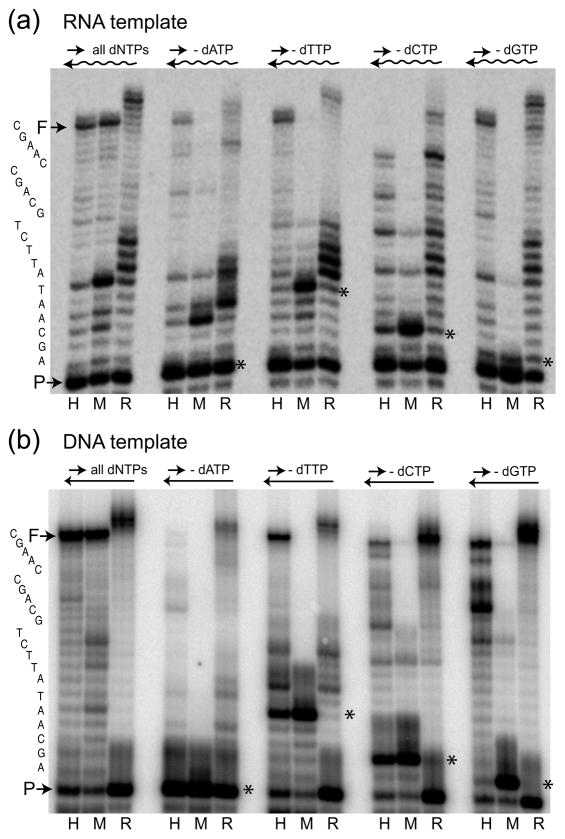
Fig. 2.
Nucleotide misincorporation assays in biased dNTP pools. The same labeled primer as in Figure 1 was annealed to a 38-mer RNA template (a) or DNA template (b) and extended by HIV-1 RT (lane H), MuLV RT (lane M) and R2 RT (lane R) at 37°C for 15 min. The experimental conditions included extension reactions with all four dNTPs (left set of lanes) and with biased dNTP pools containing only three dNTPs (− dATP, − dTTP, −dCTP or − dGTP sets of lanes). Reaction products were analyzed on a 14% polyacrylamide-urea denaturing gel. The terminal transferase activity of R2 RT generated products longer than full-length (F). The primary sequence of the correct extension product is shown on the far left. The sites for each biased pool where replication should have stopped unless misincorporations occurred are indicated with an *.