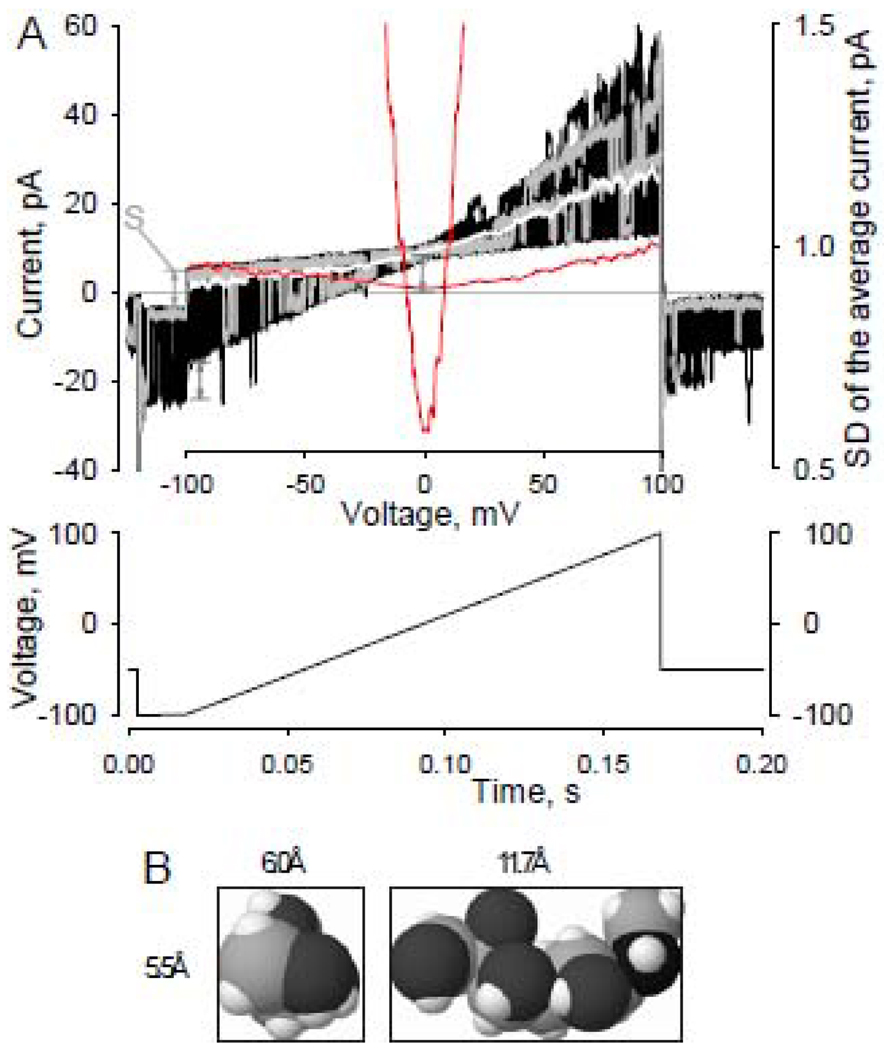
Fig. 1.
A - Patch current-voltage characteristics and reversal potential estimate. Patch current-voltage characteristics (top graph, 25 black lines, two individual traces shown in grey) were generated using series of 15-ms step at −100 mV followed by a 150-ms voltage ramp (linear change in voltage ~0.67mV/ms) from −100 mV to +100 mV were applied from a holding potential of −50 mV (diagram). The interval between sweep starts was 1s. Polarity of the protocol was changed respectively whenever the holding potential was +50mV. In order to determine the reversal potentials of unitary currents we typically used the following procedure: series of current traces (50–200 patch I–V curves) were averaged (white line) and the standard deviation (SD) for each point in the average current trace was calculated (red lines); the position of the minimum of the resulting SD curve along the voltage axis corresponds to a reversal potential value at given ionic conditions. Both red lines are the same SD of the average current function. One is scaled separately (right-hand axis) to better visualize the position of the minimum of the function. Note, absolute current values can not be used to determine Vrs. Fifteen-ms step preceding voltage ramp allows visualization of the current level shifts (S) produced by voltage ramp applications and is proportional to the uncompensated capacitance of the recording configuration and voltage ramp speed (CdV/dt). Experimental conditions: Experimental conditions: outside-out patch recording; symmetrical NaCl140 solution.
B - 3-D space-filling model representation of an NMDG molecule. Dimensions of the smallest box encompassing the molecule are 5.5Å X6.0Å X11.7Å. 6.0Å was estimated as the smallest NMDG molecule diameter. Molecular size estimate of NMDG was determined by the “smallest box enclosing solute” procedure provided by HyperChem 8.0 software (Hypercube, Inc., Gainesville, FL, USA; http://www.hyper.com).