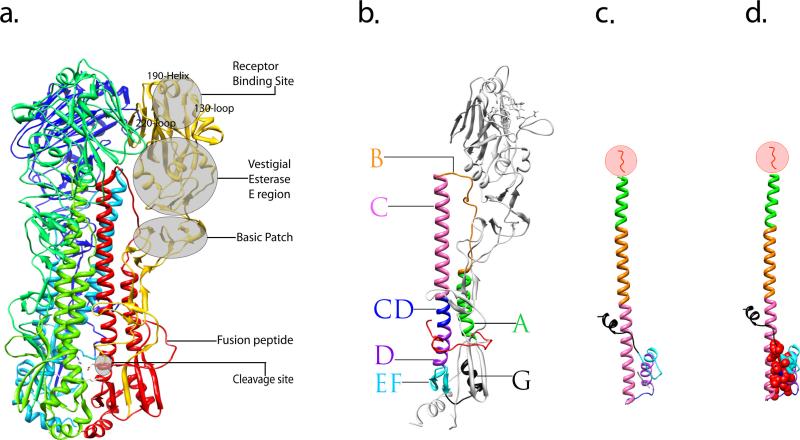
Figure 2.
Crystal structure of the influenza virus HA protein and structural characterization of its rearrangement. (a) Overview of a trimeric HA0 precursor, shown as a ribbon diagram. One monomer is colored in yellow (HA1 subunit) and red (HA2 subunit) illustrating the locations of subdomains. The two other monomers that make up the trimer are shown in blue and green. (b) Ribbon diagram of a monomeric, cleaved form of the HA. Featured subdomains are highlighted with different colors: helix A (green), loop B (orange), helix C (pink), helix CD (blue), helix D (purple), β-sheet EF (cyan), helix G (black) and fusion peptide (red). (c) Stereodiagram of the HA2 subunit in the pH induced fusion conformation, the subdomains of which correspond to that of an intact HA monomer shown in b and are indicated by the same color. (d) The stereodiagram of HA2 in panel c is shown with the residues forming the hydrophobic core marked with red spheres. In (c) and (d), the N-terminal residues 1–37 of HA2 which includes the fusion peptide (residues 1–23) were removed from during the original preparation of the protein for crystal structure [32] and is illustrated in red within the circle. Figures were prepared using Chimera [82].