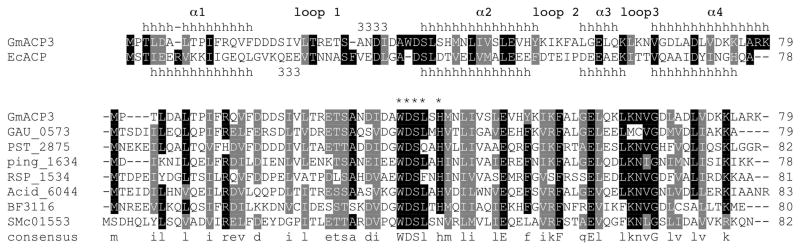
FIGURE 1.
The GmACP3 sequence family. The aligned sequences and secondary structure from E. coli ACP (from PDB ID 2FAC-A) and G. metallireducens GmACP3 are shown. The nine most similar sequences to GmACP3 reported by a BLAST search of the KEGG database (48) are aligned. KEGG IDs are given for bacterial homologs from Gemmetimonas aurantiaca (GAU_0573), Pseudomonas stutzeri (PST_2875), Psychromonas ingrahamii (Ping_1634), Rhodobacter sphaeroides (RSP_1534), Solibacter usitatus (Acid_6044), Bacteroides fragilis (BF3116), and Sinorhizobium meliloti (SMc01553). Alignments were created with ClustalW2 (71). The consensus sequence “WDSLxH/N” is indicated by asterisks (*).