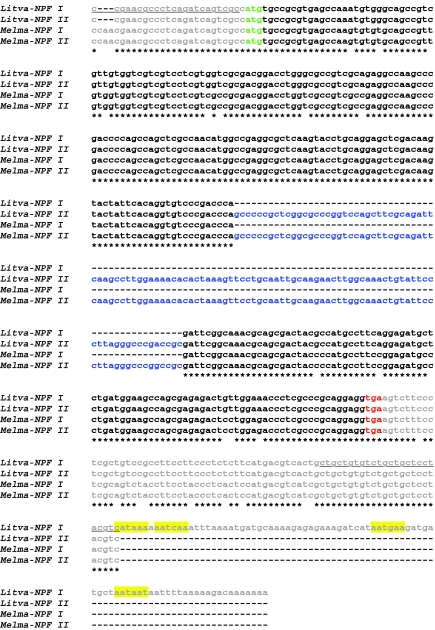
Fig. 2.
Alignment of the nucleotide sequences of Litopenaeus vannamei and Melicertus marginatus neuropeptide F-encoding transcripts. Litva-NPF I, Litopenaeus vannamei NPF I (accession no. HQ428126); Litva-NPF II; Litopenaeus vannamei NPF II (accession no. HQ428127); Melma-NPF I, Melicertus marginatus NPF I (accession no. HQ428128); Melma-NPF II, Melicertus marginatus NPF II (accession no. HQ428129). Dashes in the nucleotide sequences indicate gaps (if internal) or missing nucleotides (if at the 3′ end). Nucleotides that are shared between all four transcripts are denoted by a star. In this alignment, the 5′ and 3′ untranslated regions (UTRs) of the cDNAs are shown in gray, the start codon of each open reading frame (ORF) is shown in green and the stop codon of each transcript is shown in red (the intervening ORF sequences are shown in black, with the exception of inserts in Litva-NPF II and Melma-NPF II, which are shown in blue). Four variant polyadenylation signal sequences in the 3′ UTR of Litva-NPF I are highlighted yellow. The sequence used to design primers for RT-PCR tissue profiling is underlined.