Abstract
Background
The Culex quinquefasciatus mosquito, a major pest and vector of filariasis and arboviruses in the tropics, has developed multiple resistance mechanisms to the main insecticide classes currently available in public health. Among them, the insensitive acetylcholinesterase (ace-1R allele) is widespread worldwide and confers cross-resistance to organophosphates and carbamates. Fortunately, in an insecticide-free environment, this mutation is associated with a severe genetic cost that can affect various life history traits. Salivary proteins are directly involved in human-vector contact during biting and therefore play a key role in pathogen transmission.
Methods and Results
An original proteomic approach combining 2D-electrophoresis and mass spectrometry was adopted to compare the salivary expression profiles of two strains of C. quinquefasciatus with the same genetic background but carrying either the ace-1R resistance allele or not (wild type). Four salivary proteins were differentially expressed (>2 fold, P<0.05) in susceptible (SLAB) and resistant (SR) mosquito strains. Protein identification indicated that the D7 long form, a major salivary protein involved in blood feeding success, presented lower expression in the resistant strain than the susceptible strain. In contrast, three other proteins, including metabolic enzymes (endoplasmin, triosephosphate isomerase) were significantly over-expressed in the salivary gland of ace-1R resistant mosquitoes. A catalogue of 67 salivary proteins of C. quinquefasciatus sialotranscriptome was also identified and described.
Conclusion
The “resistance”-dependent expression of salivary proteins in mosquitoes may have considerable impact on biting behaviour and hence on the capacity to transmit parasites/viruses to humans. The behaviour of susceptible and insecticide-resistant mosquitoes in the presence of vertebrate hosts and its impact on pathogen transmission urgently requires further investigation.
Data Deposition
All proteomic data will be deposited at PRIDE (http://www.ebi.ac.uk/pride/).
Introduction
Culex pipiens quinquefasciatus is an important vector of Bancroftian filariasis and arboviruses worldwide and represents the main mosquito nuisance in urban environments [1], [2]. Vector control against this mosquito species relies essentially on environmental sanitation and the use of insecticides in polluted breeding habitats [3]. Unfortunately, resistance to insecticides in C. quinquefasciatus mosquitoes emerged more than 25 years ago in Africa, America and Europe and this resistance is frequently due to a loss of sensitivity of the insect's acetylcholinesterase enzyme to organophosphates and carbamates [4]. Two amino acid substitutions (i.e. F290V, G119S) were found to play a role in resistance [5] but the G119S resistant allele (named ace-1R) was shown to be widespread in C. quinquefasciatus natural populations [6], [7], [8], [9]. Fortunately, although insecticide resistance alleles afford a selective advantage in the presence of insecticide, they can constitute a handicap in an insecticide-free environment [10], [11]. Previous studies reported that ace-1R alleles have a strong genetic cost that can induce important behavioural and physiological changes in insects [12], [13]. In C. quinquefasciatus, ace-1 alleles coding for a modified AChE1 were associated with a longer development time, lower emergence rates and shorter wing length than in their susceptible counterparts [14], [15]. Other studies showed that the resistant larvae of C. quinquefasciatus were also less able to escape predation [16] and adult males were less competitive for mating than wild-type susceptible males [17]. The ace-1R allele is known to be the most costly of resistance genes because it interferes with the general functioning of the central nervous system throughout a mosquito's life and adversely modifies behavioural traits [18], [19].
Far less information is available about the impact of insecticide resistance alleles affecting other traits such as host seeking and blood feeding behaviour. Caroll et al. [20] first demonstrated that insecticide resistance in C. quinquefasciatus mosquitoes could interfere with the development of parasites, i.e. organophosphate-resistant mosquitoes were less likely to transmit filariasis than their insecticide-susceptible counterparts [20]. Before taking a blood meal, mosquito females inject several salivary substances into the host skin to counteract the haemostatic reaction induced by the bite [21]. The main functions of the saliva are powerful anti-coagulation, vasodilatation and platelet aggregation inhibition [22] that favour the blood feeding success. Mosquito salivary proteins then play a major role in host-vector interaction and can also interfere with pathogen transmission [23] including that of arboviral viruses [24] and parasites [25]. Here, an original approach by proteomic technology combining 2D-electrophoresis (2DE) and mass-spectrometry (MS) was used to compare the salivary expression profile of two strains of C. quinquefasciatus having same genetic background but either carrying the ace-1R resistance allele or not (wild type). The hypothesis is that the genetic cost associated with the ace.1R allele may modulate the expression of salivary proteins in C. quinquefasciatus salivary glands. In our study, differences between strains could be directly attributed to the expression of resistance alleles in a standard genetic environment. An updated list of salivary proteins from C. quinquefasciatus sialotranscriptome is also provided.
Materials and Methods
Culex mosquito strains
Two strains of C. quinquefasciatus were used; SLAB and SR. They all share the same genetic background and only differ in their genotype at the ace-1 locus [15]. SLAB, the insecticide-susceptible reference strain [26], is homozygous for susceptible alleles at the ace-1 locus. SR is homozygous for the resistant allele ace-1R which was introgressed into the genome of SLAB through 14 repeated generations of backcrossing [17].
Preparation of salivary gland extracts
Unfed mosquitoes of the SLAB and SR strains, 7 days old, were first sedated with CO2. The salivary glands were dissected and then transferred to rehydratation buffer and stored at −80°C before use. A total of 30 batches of 30 pairs of salivary glands from C. quinquefasciatus females were obtained per strain. The salivary glands were lysed in liquid nitrogen and homogenates were then centrifuged for 30 min at 30,000 g at 4°C. The supernatants, named Salivary Gland Extracts (SGE) containing soluble salivary proteins, were subjected to two-dimensional electrophoresis (2DE). All reagents used for 2DE were from the Plus One range (GE Healthcare Biosciences, Uppsala, Sweden).
Two-dimensional electrophoresis
2DE was carried out with 22 µg of C. quinquesfasciatus SGE on 11 cm immobiline™ dryStrips pH 3–11 non linear (NL) (GE Healthcare, Germany). Strips were rehydrated for 10–20 h at 20°C with protein samples made up to 170 μl by adding IEF buffer (7 M urea, 2 M thiourea, 4% CHAPS, 0.2% tergitol, 0.8% IPG buffer, and 1,2% DeStreak reagent). Running conditions were: temperature 20°C; current 50 μA per strip; 300 V (gradient) for 5 min; 300 V (step) for 30 min, 5000 V (gradient) for 3 h, and then 5000 V steps up to 60000 Vh. The second dimension was carried out on 10–20% SDS-PAGE gels (Biorad, Marnes la Coquette, France) at 70 V for 15 min and then 200 V until the bromophenol blue front had reached the end of the gel. The gels were fixed 20 minutes in 50% ethanol/5% acetic acid solution, and then 10 min in 50% ethanol solution, washed four times in MilliQ water. Finally, gels were stained with colloidal coomassie blue (Fermentas, Saint-Remy les Chevreuse, France) overnight and washed twice in milliQ water. Gels were scanned with a EPSON Perfection pro V750. All gel images were acquired at 16 bits resolution under non saturating conditions. 2DE images were analyzed using Same Spots™ Software 3.3 (Nonlinear Dynamics). Statistical analysis and protein quantification were carried out using the same software. First, PCA analysis was performed to verify that the gels from both strains (SLAB and SR) were distributed in two distinct groups. Secondly, statistical analysis was done by an ANOVA test (P<0.05) for all spots in both groups. About 20 spots reached the threshold of significant differential expression between both strains, and a second statistical analysis taking into account possible false positives was then performed with a cut-off of 2 fold in either direction (up and down-expression) and with P<0.05 and power >0.8. The q value represents therefore the P value adjusted by the False Discovery Rate (FDR). Details are indicated in: http://www.nonlinear.com/support/progenesis/samespots/faq/pq-values.aspx). Protein spots of each strain were digested by trypsin and identified by mass-spectrometry.
Identification of salivary proteins by mass-spectrometry
Trypsin digestion
Enzymatic in-gel digestion was performed automatically (Tecan freedom evo® proteomics) according to the Shevchenko modified protocol [27].
Briefly, protein spots were digested using 150 ng of trypsin, peptide extraction was performed using 5 sonication cycles of 2 min each and peptides were concentrated 1 hour at 50°C in a heat block. Peptide samples were automatically spotted (Tecan freedom evo® proteomics). For this step, 0.5 µl of sample peptide and 0.5 µl of alpha-cyano-4-hydroxy-trans-cinnamic acid (a saturated solution prepared in acetonitrile/trifluoroacetic acid, 50 ∶ 0.1%, vortexed, sonicated 30 s and microcentrifuged 30 s with a 1/3 dilution of the supernatant used as the matrix) were deposited on a 384-well MALDI anchorship target using the dry-droplet procedure [28] and air dried at room temperature. Peptide samples were then desalted using a 10 mM phosphate buffer and dried again at room temperature. Mass spectrometry was then performed on both SLAB and SR strains of C. quinquefasciatus.
MALDI-TOF MS analysis
Analyses were performed using an UltraFlex MALDI TOF-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) in the reflectron mode with a 26 kV accelerating voltage and a 50 ns delayed extraction. Mass spectra were acquired in automatic mode using the AutoXecute™ module of Flexcontrol™ (Bruker Daltonics) (laser power ranged from 40 to 50%, 600 shots). Spectra were analyzed using FlexAnalysis™ software (Bruker Daltonics) and calibrated internally with the autoproteolysis peptides of trypsin (m/z: 842.51; 1045.56; 2211.10). Peptides were selected in the mass range of 900–3000 Da.
Peptide mass fingerprint identification of proteins was performed by searching against the Insecta entries of either SwissProt or TrEMBL databases (http://www.expasy.ch) using the Mascot v 2.2 algorithm (http://www.matrixscience.com) as previously described [29] Mascot scores higher than 65 were considered as significant (P<0.05) for the SwissProt and TrEMBL databases
Nano LC-MS/MS analysis
Protein samples that could not be identified by MALDI-TOF MS analysis were subjected to nano LC ESI MS/MS analysis with a QTOF or a LTQ Orbitrap XL. Samples were dehydrated in a vacuum centrifuge, solubilized in 1 µl of 0.1% formic acid-2% acetonitrile and analyzed online on a ESI quadrupole time-of-flight (Q-TOF) mass spectrometer (QSTAR Pulsar-i, Applied Biosystems, Foster City, CA) or a ESI LTQ Orbitrap mass spectrometer (LTQ Orbitrap XL, Thermo Fisher Scientific) respectively, coupled with an Ultimate 3000 HPLC (Dionex, Amsterdam, Netherlands). Details are given in Table S1.
Regarding the protein identification, all MS/MS spectra were searched against the Insecta entries of either SwissProt or TrEMBL databases by using the Mascot v 2.2 algorithm (MA; Matrix Science Inc.) with trypsin enzyme specificity and one missed trypsin cleavage. With nano LC ESI LTQ Orbitrap XL analysis, the data submission was performed using ProteomeDiscoverer v 1.0 (Thermo Fisher Scientific). Peptides with scores greater than the identity score (P<0.05) were considered significant. All spectra were manually validated for proteins identified with less than three different peptides.
Results
Differential expression profile of sialome between susceptible and ace-1R resistant Culex mosquitoes
Differential sialome expression between susceptible (SLAB) and resistant (SR) strains of C. quinquefasciatus was assessed by comparing 2D-electrophoresis gels.
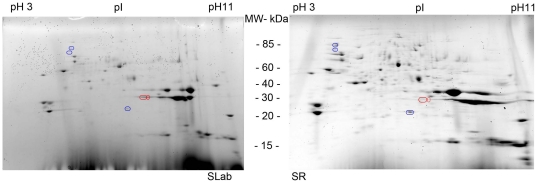
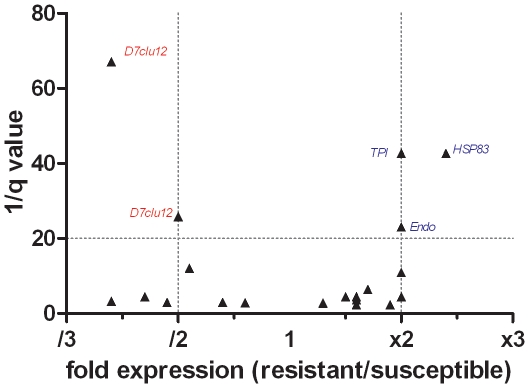
In overall, 14 gels were obtained for each strain (SLAB and SR) and 322 spots (excluding artefacts) were detected (Figure 1). Six of 14 gels were excluded per strain because of unreliable spot focalization. On the remaining gels, Principal Component Analysis showed that spot profiles significantly differed between the resistant and susceptible strains (variance >46%, data not shown). The first set of ANOVA analysis detected 20 spots showing differential expression between the SLAB and SR strains (Figure 2) but after adjustment using the FDR approach, only 5 of these showed significant differential expression (q<0.05, Power >0.8) (Figure 2, Table S1). Two spots (in red) showed two-fold under-expression in resistant strain SR compared to the SLAB strain whereas three other spots (in blue) showed two-fold over-expression in the resistant strain.
Figure 1. 2D electrophoresis profile (SDS-Page) of C. quinquefasciatus salivary gland proteins.
Salivary gland extracts (SGE) from the susceptible SLAB strain (left panel) and the ace.1 resistant SR strain (right panel) are shown. Concentrations were measured according to the Bradford method; 729 µg/ml for SLAB strain and 816 µg/ml for SR strain. Protein spots showing increased or decreased expression in the resistant strain are circled in blue and red, respectively.
Figure 2. Differential salivary protein expression between susceptible and ace.1 resistant C. quinquefasciatus.
Salivary gland extractions were carried out using 7-day old unfed females. Differences in protein expression are indicated as a function of both expression ratio (resistant/susceptible) and significance ratio (q value). Vertical lines indicate two-fold differential expression in either direction. Horizontal lines indicate the significance threshold (1/q<20) or q<0.05) according to Same Spot analysis. Proteins showing more than 2.0 fold expression and a significant 1/q value are named.
Identification of salivary gland proteins by mass spectrometry
The second step was to characterize the sialome of C. quinquefasciatus using both MALDI-TOF and LC-MS/MS mass-spectrometry. A total of 89 spots were analyzed by MA and a catalogue of 52 salivary proteins were described (Figure 3). The identified proteins could be split into three functional classes: “salivary” products, housekeeping products and products of “unknown” function (Table 1). Of the identified proteins, 43% were secreted and 57% were intracellular.
Figure 3. Identification of C. quinquefasciatus salivary proteins by mass spectrometry.
Proteins are indicated in a 2D-gels for susceptible and resistant (ace.1R) mosquitoes with spots named according to Table 1. A total of 89 spots were analyzed and a catalogue of 52 salivary proteins is shown. Proteins which are more highly expressed in the resistant (SR) strain than SLAB are coded in blue whereas those under-expressed are coded in red. Spots were excised manually and then digested and extracted with TECAN EVO or manually. Proteins were identified by MALDI-TOF and if the identification was ambiguous, LC-MS/ MS was performed.
Table 1. Identification and classification of C. quinquefasciatus salivary proteins.
| Abbreviation | Protein Identification | function | Accession Number | Nominal Mass (kDa) | Pi | Sequence Coverage % | MS source | Spot ID |
| NADPH2 | Glutamate semialdehyde dehydrogenase – Culex quinquefasciatus | amine metabolism | B0X119_CULQU | 86226 | 6,7 | 36 | MALDI TOF | 545–606 |
| 30 kDa Cq | 30 kDa salivary gland allergen Aed a 3 – Culex quinquefasciatus | antigen V family | B0W7N1_CULQU | 27712 | 4,57 | 5 | LC-MS/MS QTOF | 521–565 522 |
| AG5-3 | Salivary secreted antigen-5 AG5-3 – Culex quinquefasciatus | antigen V family | B0XGB4_CULQU | 29925 | 7,14 | 4 | LC-MS/MS QTOF | 558 |
| ADA | Salivary adenosine deaminase – Culex quinquefasciatus | blood feeding | Q95WT8_CULQU | 57849 | 5,86 | 32 | MALDI TOF | 541–544–573 |
| Apy | Apyrase – Culex quinquefasciatus | blood feeding | B0WUA1_CULQU | 59481 | 7,21 | 21 | MALDI TOF | 597 |
| D7 Clu 1 | Long form D7clu1 salivary protein – Culex quinquefasciatus | blood feeding | Q95V93_CULQU | 35750 | 7,45 | 41 | MALDI TOF | 553–555–578–580 |
| D7 Clu 12 | Long form D7clu12 salivary protein – Culex quinquefasciatus | blood feeding | Q95V92_CULQU | 36537 | 7,5 | 51 | MALDI TOF | 581–558–523–530–564–587–588 |
| C = D7-L3 | Salivary long D7 protein 3 – Culex quinquefasciatus | blood feeding | B0X6Z3_CULQU | 27332 | 7,53 | 15 | LTQ-Orbitrap | 594 = C |
| D7-L3 | Salivary long D7 protein 3 – Culex quinquefasciatus | blood feeding | Q6TRZ6_CULQU | 39205 | 6,67 | 29 | MALDI TOF | 579 |
| 5-Nu | Salivary apyrase; 5′ nucleotidase – Culex quinquefasciatus | blood feeding | B0XHG2_CULQU | 62553 | 5,47 | 21 | MALDI TOF | 602 |
| E3 | Dihydrolipoyl dehydrogenase, mitochondrial – Culex quinquefasciatus | citric acid cycle, glycolyis | B0X2P1_CULQU | 37768 | 5,53 | 38 | MALDI TOF | 547 = A, 549,607 |
| ALDH | Aldehyde dehydrogenase, mitochondrial – Culex quinquefasciatus | B0WKSO_CULQU | 57133 | 7,64 | 23 | |||
| E2 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase Culex quinquefasciatus | B0XAPO_CULQU | 54962 | 9 | 22 | |||
| SCSb | Succinyl-coa synthetase beta chain – Culex quinquefasciatus | citric acid cycle | B0WFW7_CULQU | 48729 | 7,1 | 16 | LTQ-Orbitrap | 594 = C |
| AcoA | Acyl-coa dehydrogenase – Culex quinquefasciatus | citric acid cycle | B0WLI6_CULQU | 46057 | 8,37 | 10 | LTQ-Orbitrap | 594 = C |
| MLC | Myosin light chain 2 – Culex quinquefasciatus | cytosqueletton | B0XDR8_CULQU | 22838 | 4,65 | 29 | LC-MS/MS QTOF | 521–565 |
| VTG | Vitellogenin – Culex quinquefasciatus | endocrinal pathway | B0X7X6_CULQU | 42128 | 8,36 | 11 | MALDI TOF | 585 |
| HADHB | 3-ketoacyl-CoA thiolase – Culex quinquefasciatus | fatty acid oxydation | B0W5M7_CULQU | 41730 | 8,59 | 2 | MALDI TOF | 585 |
| MDH | Malate dehydrogenase – Culex quinquefasciatus | gluconeogenesis | B0W5T5_CULQU | 35315 | 6,15 | 32 | MALDI TOF | 574–542 |
| PC | Pyruvate carboxylase, mitochondrial – Culex quinquefasciatus | gluconeogenesis | B0W649_CULQU | 133344 | 6,59 | 34 | MALDI TOF | 540 |
| ENO | Enolase – Culex quinquefasciatus | glycolysis | B0W1N4_CULQU | 46908 | 6,29 | 19 | MALDI TOF | 550 |
| TPI | Triosephosphate isomerase – Culex quinquefasciatus | glycolysis | B0W5W4_CULQU | 24075 | 6 | 12 | LC-MS/MS QTOF | 543 |
| C = CAP | Adenylyl cyclase-associated protein – Culex quinquefasciatus | inositol cycle | B0W727_CULQU | 68046 | 5,32 | 10 | LTQ-Orbitrap | 594 = C |
| DNAse 1 | Deoxyribonuclease I – Culex quinquefasciatus | nucleic acid metabolism | B0W7Z5_CULQU | 46863 | 5,61 | 20 | MALDI TOF | 593 |
| Enase | CULQU Salivary endonuclease – Culex quinquefasciatus | nucleic acid metabolism | B0WQ10_CULQU | 42721 | 9,28 | 3 | MALDI TOF | 585 |
| CALR | Calreticulin – Culex quinquefasciatus | protein folding | B0WJE0_CULQU | 46874 | 4,37 | 51 | MALDI TOF | 596– 524 |
| PDI-1 | Disulfide isomerase – Culex quinquefasciatus | protein folding | B0X3M7_CULQU | 55733 | 4,82 | 46 | MALDI TOF | 525–566 |
| PDI- 2 | Disulfide isomerase – Culex quinquefasciatus | protein folding | B0X904_CULQU | 54158 | 5,9 | 40 | MALDI TOF | 534–572–595 |
| Endo | Endoplasmin – Culex quinquefasciatus | protein folding | B0W5Z4_CULQU | 91045 | 4,9 | 40 | MALDI TOF | 592–526–591 |
| HSP 70 | Heat shock 70 kDa protein cognate 4 – Culex quinquefasciatus | protein folding- stress response | B0WP93_CULQU | 71787 | 5,36 | 41 | MALDI TOF | 532–609 |
| HSP 83 | Heat shock protein 83 – Culex quinquefasciatus | protein folding- stress response | B0WX04_CULQU | 81938 | 4,9 | 37 | MALDI TOF | 590 |
| EF1a | Elongation factor 1-alpha 1 – Culex quinquefasciatus | protein synthesis | B0WQ61_CULQU | 52582 | 9,23 | 29 | MALDI TOF | 559 |
| EF1B | Elongation factor 1-beta – Culex quinquefasciatus | protein synthesis | B0WF81_CULQU | 24630 | 4,57 | 9 | LC- MS/MS QTOF | 521–565 |
| EF 2 | Elongation factor 2 – Culex quinquefasciatus | protein synthesis | B0W238_CULQU | 115611 | 6,29 | 19 | MALDI TOF | 546 |
| SAF | Spermatogenesis associated factor – Culex quinquefasciatus | related to ATP-binding proteins | B0WC89_CULQU | 88819 | 5,23 | 18 | MALDI TOF | 604 |
| GRP 78 | 78 kDa glucose-regulated protein - Culex quinquefasciatus | related to HSP 70 family | B0W934_CULQU | 72377 | 5,07 | 37 | MALDI TOF | 605 |
| Punp | Putative uncharacterized protein- Culex quinquefasciatus | related to HSP family | B0WKR8_CULQU | 103860 | 4,93 | 26 | MALDI TOF | 600 |
| CytC | Ubiquinol-cytochrome c reductase complex core protein - Culex quinquefasciatus | respiratory reaction | B0WXM0_CULQU | 45651 | 9,02 | 11 | MALDI TOF | 585 |
| C = Hrb27c | Heterogeneous nuclear ribonucleoprotein 27C OS - Culex quinquefasciatus | ribonucleosome component | B0X7P8_CULQU | 42660 | 6,54 | 18 | LTQ-Orbitrap | 594 = C |
| GAA | Alpha-glucosidase - Culex quinquefasciatus | sugar feeding | B0XAA1_CULQU | 66690 | 5,08 | 36 | MALDI TOF | 567–610 |
| PYG | Glycogen phosphorylase - Culex quinquefasciatus | sugar metabolism | B0WCF2_CULQU | 97096 | 5,96 | 22 | MALDI TOF | 539 |
| 15.8p | Putative 15.8 kDa salivary peptide - Culex quinquefasciatus | unknown | Q6TRX5_CULQU | 17826 | 8,75 | 39 | MALDI TOF | 598 |
| 16.4p | 16.4 kDa salivary peptide - Culex quinquefasciatus | unknown | Q6TS05_CULQU | 18312 | 6,82 | 37 | MALDI TOF | 556–582 |
| 20.2p | 20.2 kDa salivary peptide - Culex quinquefasciatus | unknown | B0WZL9_CULQU | 23264 | 8,65 | 39 | MALDI TOF | 557–583–599 |
| 16p | 16 kDa salivary peptide - Culex quinquefasciatus | unknown | Q6TRZ5_CULQU | 18301 | 9,02 | 15 | LC-MS/MS QTOF | 584 = B |
| 16.8p | Putative 16.8 kDa salivary protein - Culex quinquefasciatus | unknown | Q6TRY2_CULQU | 18773 | 8,66 | 7 | ||
| 16.8p1 | Putative 16.8 kDa salivary peptide - Culex quinquefasciatus | unknown | Q6TS03_CULQU | 19267 | 8,78 | 6 | ||
| H2BF | Histone H2B.a/g/k - Culex quinquefasciatus | DNA structure | B0WG57_CULQU | 5744 | 10,1 | 16 | ||
| 13.1p | Putative 13.1 kDa salivary protein - Culex quinquefasciatus | unknown | Q6TS27_CULQU | 15209 | 9,47 | 8 | ||
| Punp1 | Putative uncharacterized protein - Culex quinquefasciatus | unknown | B0X1P5_CULQU | 27764 | 4,72 | 23 | LC-MS/MS QTOF | 522 |
| SP | Salivary protein - Culex quinquefasciatus | unknown | B0W4E3_CULQU | 24461 | 4,54 | 7 | LC-MS/MS QTOF | 521–565 |
| SP2 | Salivary protein - Culex quinquefasciatus | unknown | Q6TS09_CULQU | 23785 | 9,18 | 67 | MALDI TOF | 562 |
Database searches used the MASCOT program and SwissProt/TrEMBL databases. Molecular mass, pI and sequence coverage are shown. All the MASCOT scores are p>65. Proteins are divided into housekeeping products (proteins expressed routinely in the cell) salivary proteins, including proteins involved in blood and/or sugar feeding, and unknown proteins for which no function has been defined.
Among the “salivary products” that are specifically expressed in the mosquito salivary glands, two spots corresponding to a unique protein named the D7 long form cluster12 protein showed two-fold lower expression in the resistant strain SR compared to the SLAB strain (Figure 2). The D7 family is widely distributed in mosquito salivary glands and plays a key role in blood feeding success [30], [31]. Other salivary proteins were identified including apyrase, 5′nucleotidase, antigen 5 family (30 kDa Cq, AG5-3) and adenosine deaminase, which is believed to be involved in blood feeding (Figure 3, Table 1).
Of the housekeeping products, which include many proteins involved in metabolism, three proteins showed two-fold higher expression in the resistant strain (Figure 2). These proteins were identified as endoplasmin, triosephosphate isomerase and a heat shock protein (HSP83). These proteins are known to be involved in protein folding, glycolysis and stress response, respectively. Among other housekeeping proteins, the salivary endonuclease which belongs to the hydrolase family was also identified (Table 1): this protein is believed to reduce local blood viscosity at the bite site to enhance the feeding process [31].
Among the “unknown” products, no significant difference in protein expression was noted between the susceptible and resistant strains (P>0.05). Most of secreted salivary peptides identified belonged to the cystein and tryptophan rich protein family of the Culex genus (CWRP- peptide 15.8p, 16.4p, 16p, 16.8p, 16.8p1, and 13.1p). As previously described, this family is highly expressed in the salivary glands of C. quinquesfacsiatus [31]: it has been proposed that these may antagonise serotonin and histamine but their role in the host-vector relationships needs further investigation.
Discussion
In the present study, we compared the expression of salivary proteins of two mosquito strains of C. quinquefasciatus with same genetic background but carrying either the ace-1R resistance allele or not.
Our results showed that four proteins were differentially expressed in the C. quinquefasciatus sialome between the resistant (ace.1 R) and the susceptible strain (q<0.05 and Power >0.8). To our knowledge, this is the first evidence that an insecticide-resistance gene can modulate the expression of salivary proteins in Diptera. Among these 4 proteins, the D7 long form salivary protein, which is secreted by mosquitoes during the blood meal [32], was significantly underexpressed in the resistant strain compared to the susceptible strain. This protein is present in all haematophagous diptera and may play a major role in blood feeding success. In mosquitoes, the D7 long form protein is an important component of the salivary glands and belongs to the super family of odorant binding proteins [30]. Although their function in blood feeding remains unclear, it has been suggested that this protein could sequester and inhibit biogenic amines (serotonin, histamine) involved in inflammation and pain during the bite [31]. The consequences of lower expression of the D7 protein are currently unknown but it speculates that its down-expression may compromise C. quinquefasciatus blood feeding and could therefore affect the transmission of pathogens to humans. In addition, major salivary proteins - including the apyrase and D7 proteins - are known to be significantly down-regulated in infected Anopheles compared to uninfected mosquitoes [33], [34]. These findings emphasise the need for further experiments to assess the impact of pathogen infection on salivary protein expression in both susceptible and Ace.1R resistant mosquitoes.
Conversely, three other proteins, namely endoplasmin, triosephosphate isomerase and heat shock protein (HSP) were significantly over-expressed in the salivary glands of Ace.1 resistant mosquitoes. Triosephosphate isomerase is an enzyme involved in glycolysis which takes place in the cytosol of cells. Over-expression of this enzyme suggests modulation of the metabolic activity of the salivary gland in the resistant strain. The HSP was the most stress-responsive protein in diptera and has also been identified in humans and rodents [35]. Over-expression of this protein may be explained by the fact that salivary gland tissues may be modified in the presence of Ace.1 R allele, e.g. by stress. Endoplasmin is produced by the endoplasmic reticulum and acts as molecular chaperone to transport secreted proteins. Its over-expression in resistant mosquitoes may suggest modulation of salivary gland cells due to the presence of the acetylcholinesterase resistance gene.
The present study also provided important data on the composition of the salivary gland of C. quinquefasciatus. Most of the identified proteins (57%) are involved in energy pathways, sugar feeding, protein folding, and the stress response. The remaining 43% are secreted proteins possibly associated with the blood meal. This finding confirms the work of Ribeiro et al [30] who reported a similar proportion of secreted proteins in Culex mosquitoes at the transcriptional level.
Previous proteomic studies showed that the secreted D7 proteins occurred in five short forms and two long forms in Anopheles mosquitoes [36] whereas two short forms and two long forms have been described in Aedes [37]. In C. quinquefasciatus, we found only three long forms of D7 (D7 Cluster12, D7 Cluster1, D7L3) which is inconsistent with previous results [30]. This may be explained by a failure to detect a small numbers of transcripts for short forms using a proteomic approach. Finally, salivary peptides (including those of the CWRP family) were identified by mass-spectrometry but the role of these proteins in Culex salivary glands requires further investigation.
Previous studies have demonstrated that the presence of the Ace.1R allele can affect several life history traits in mosquitoes [15], [17] Djogbénou et al, [38] recently reported a lack of Ace.1R homozygous resistance in An. gambiae populations from Burkina Faso, suggesting that the mutation may have a significant genetic cost. Effects of the Ace.1R allele on salivary protein down-expression might affect the fitness of homozygous resistant mosquitoes if their blood feeding success is significantly compromised by the presence of the mutation. Salivary proteins are injected into the skin to counteract the host's haemostatic reaction to the bite [39] and any modification in saliva composition could then modify the host's haemostatic response and thereby interfere with blood feeding success. Behavioural investigations using a video tracking system are needed to compare the flying, probing and biting behaviour of insecticide-resistant and susceptible mosquitoes. This will shed light on the impact of the modification of salivary proteins on the global fitness of resistant mosquitoes.
Supporting Information
Salivary protein expression data.
(DOC)
Acknowledgments
We thank Marie Noelle Lacroix for help on mosquito rearing in Montpellier (LIN/IRD).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Innocent Djegbe was supported by a fellowship provided by the IRD (Département Soutien et Formation). This research was supported by the French Institut de Recherche pour le Développement (IRD) and the Anopheles Biology & Control (ABC) network. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Maxwell CA, Mohammed K, Kisumku U, Curtis CF. Can vector control play a useful supplementary role against bancroftian filariasis? Bull World Health Organ. 1999;77:138143. [PMC free article] [PubMed] [Google Scholar]
- 2.Dohm DJ, O'Guinn ML, Turell M. Effect of environmental temperature on the ability of Culex pipiens (Diptera: Culicidae) to transmit West Nile virus. J Med Entomol. 2002;39:221–225. doi: 10.1603/0022-2585-39.1.221. [DOI] [PubMed] [Google Scholar]
- 3.WHO. Guidelines for testing mosquito adulticides intended for Indoor Residual Spraying (IRS) and Insecticide Treated Nets (ITNs). 2006. WHO/CDS/NTD/WHOPES/GCDDP 3. [DOI] [PMC free article] [PubMed]
- 4.Lenormand T, Bourguet D, Guillemaud T, Raymond M. Tracking the evolution of insecticide resistance in the mosquito Culex pipiens. Nature London. 1999;401:861–864. doi: 10.1038/23685. [DOI] [PubMed] [Google Scholar]
- 5.Alout H, Labbe P, Berthomieu A, Pasteur N, Weill M. Multiple duplications of the rare ace-1 mutation F290V in Culex pipiens natural populations. Insect Biochem Mol Biol. 2009 doi: 10.1016/j.ibmb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Weill M, Duron O, Labbe P, Berthomieu A, Raymond M. [Insecticide resistance in the mosquito Culex pipiens]. Med Sci (Paris) 2003;19:1190–1192. doi: 10.1051/medsci/200319121190. [DOI] [PubMed] [Google Scholar]
- 7.Cui F, Raymond M, Berthomieu A, Alout H, Weill M, et al. Recent emergence of insensitive acetylcholinesterase in Chinese populations of the mosquito Culex pipiens (Diptera: Culicidae). J Med Entomol. 2006;43:878–883. doi: 10.1603/0022-2585(2006)43[878:reoiai]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Labbe P, Berthomieu A, Berticat C, Alout H, Raymond M, et al. Independent duplications of the acetylcholinesterase gene conferring insecticide resistance in the mosquito Culex pipiens. Mol Biol Evol. 2007;24:1056–1067. doi: 10.1093/molbev/msm025. [DOI] [PubMed] [Google Scholar]
- 9.Tantely L, Tortosa P, Alout H, Berticat C, Berthomieu A, et al. Insecticide resistance in Culex pipiens quinquefasciatus and Aedes albopictus mosquitoes from La Réunion Island. Insect Bioch Mol Biol. 2010;40:317–324. doi: 10.1016/j.ibmb.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Raymond M, Berticat C, Weill M, Pasteur N, Chevillon C. Insecticide resistance in the mosquito Culex pipiens: what have we learned about adaptation? Genetica. 2001;112–113:287–296. [PubMed] [Google Scholar]
- 11.Agnew P, Berticat C, Bedhomme S, Sidobre C, Michalakis Y. Parasitism increases and decreases the costs of insecticide resistance in mosquitoes. Evolution Int J Org Evolution. 2004;58:579–586. [PubMed] [Google Scholar]
- 12.Weill M, Labbé P, Duron O, Pasteur N, Fort P, et al. Insecticide resistance in the mosquito Culex pipiens: towards an understanding of the evolution of ace genes. Insect Evolutionary Ecology 2005 [Google Scholar]
- 13.Djogbenou L, Noel V, Agnew P. Costs of insensitive acetylcholinesterase insecticide resistance for the malaria vector Anopheles gambiae homozygous for the G119S mutation. Malar J. 2010;9:12. doi: 10.1186/1475-2875-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourguet D, Guillemaud T, Chevillon C, Raymond M. Fitness costs of insecticide resistance in natural breeding sites of the mosquito Culex pipiens. Evolution Int J Org Evolution. 2004;58:128–135. doi: 10.1111/j.0014-3820.2004.tb01579.x. [DOI] [PubMed] [Google Scholar]
- 15.Berticat C, Bonnet J, Duchon S, Agnew P, Weill M, et al. Costs and benefits of multiple resistance to insecticides for Culex quinquefasciatus mosquitoes. BMC Evol Biol. 2008;8:104. doi: 10.1186/1471-2148-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berticat C, Duron O, Heyse D, Raymond M. Insecticide resistance genes confer a predation cost on mosquitoes, Culex pipiens. Genet Res. 2004;83:189–196. doi: 10.1017/s0016672304006792. [DOI] [PubMed] [Google Scholar]
- 17.Berticat C, Boquien G, Raymond M, Chevillon C. Insecticide resistance genes induce a mating competition cost in Culex pipiens mosquitoes. Genet Res. 2002;79:41–47. doi: 10.1017/s001667230100547x. [DOI] [PubMed] [Google Scholar]
- 18.Shi MA, Lougarre A, Alies C, Fremaux I, Tang ZH, et al. Acetylcholinesterase alterations reveal the fitness cost of mutations conferring insecticide resistance. BMC Evol Biol. 2004;4:5. doi: 10.1186/1471-2148-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourguet D, Lenormand T, Guillemaud T, Marcel V, Fournier D, et al. Variation of dominance of newly arisen adaptive genes. Genetics. 1997;147:1225–1234. doi: 10.1093/genetics/147.3.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarroll L, Paton MG, Karunaratne SH, Jayasuryia HT, Kalpage KS, et al. Insecticides and mosquito-borne disease. Nature. 2000;407:961–962. doi: 10.1038/35039671. [DOI] [PubMed] [Google Scholar]
- 21.Ribeiro JM, Francischetti I. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro JM. Blood-feeding in mosquitoes: probing time and salivary gland anti-haemostatic activities in representatives of three genera (Aedes, Anopheles, Culex). Med Vet Entomol. 2000;14:142–148. doi: 10.1046/j.1365-2915.2000.00227.x. [DOI] [PubMed] [Google Scholar]
- 23.Edwards JF, Higgs S, Beaty BJ. Mosquito feeding-induced enhancement of Cache Valley Virus (Bunyaviridae) infection in mice. J Med Entomol. 1998;35:261–265. doi: 10.1093/jmedent/35.3.261. [DOI] [PubMed] [Google Scholar]
- 24.Schneider BS, Higgs S. The enhancement of arbovirus transmission and disease by mosquito saliva is associated with modulation of the host immune response. Trans R Soc Trop Med Hyg. 2008;102:400–408. doi: 10.1016/j.trstmh.2008.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donovan MJ, Messmore AS, Scrafford DA, Sacks DL, Kamhawi S, et al. Uninfected mosquito bites confer protection against infection with malaria parasites. Infect Immun. 2007;75:2523–2530. doi: 10.1128/IAI.01928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georghiou GP, Metcalf RL, Gidden FE. Carbamate-resistance in mosquitos. Selection of Culex pipiens fatigans Wiedemann ( = C. quinquefasciatus Say) for resistance to Baygon. Bull World Health Organ. 1966;35:691–708. [PMC free article] [PubMed] [Google Scholar]
- 27.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 28.Karas M, Hillenkamp F. Laser desorption ionization of protein with molecular mass exceeding 10,000 daltons. Anal chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 29.Wasinpiyamongkol L, Patramool S, Luplertlop N, Surasombatpattana P, Doucoure S, et al. Blood-feeding and immunogenic Aedes aegypti saliva proteins. Proteomics. 2010;10:1906–1916. doi: 10.1002/pmic.200900626. [DOI] [PubMed] [Google Scholar]
- 30.Ribeiro J, Charlab R, Pham V, Garfield M, Valenzuela J. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34:543–563. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Calvo E, Pham VM, Lombardo F, Arca B, Ribeiro JM. The sialotranscriptome of adult male Anopheles gambiae mosquitoes. Insect Biochem Mol Biol. 2006;36:570–575. doi: 10.1016/j.ibmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Nascimento E, dos Santos Malafronte R, Marinotti O. Salivary gland proteins of the mosquito Culex quinquefasciatus. Arch Insect Biochem Physiol. 2000;43:9–15. doi: 10.1002/(SICI)1520-6327(200001)43:1<9::AID-ARCH2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Rossignol PA, Ribeiro JM, Spielman A. Increased intradermal probing time in sporozoite-infected mosquitoes. Am J Trop Med Hyg. 1984;33:17–20. doi: 10.4269/ajtmh.1984.33.17. [DOI] [PubMed] [Google Scholar]
- 34.Choumet V, Carmi-Leroy A, Laurent C, Lenormand P, Rousselle J, et al. The salivary glands and saliva of Anopheles gambiae as an essential step in the Plasmodium life cycle: a global proteomic study. Proteomics. 2007;7:3384–3394. doi: 10.1002/pmic.200700334. [DOI] [PubMed] [Google Scholar]
- 35.Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, et al. Déjà vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8:1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 36.Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, et al. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- 37.Ribeiro JM, Arca B, Lombardo F, Calvo E. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djogbenou L, Chandre F, Berthomieu A, Dabire R, Koffi A, et al. Evidence of introgression of the ace-1(R) mutation and of the ace-1 duplication in West African Anopheles gambiae s. s. PLoS ONE. 2008;3:e2172. doi: 10.1371/journal.pone.0002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Champagne DE, Valenzuela JG. Pharmacology of haematophagous arthropod saliva; The Immunology of host-ectoparasitic arthropod relationships. Wallingford: CAB international; 1996. pp. 85–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Salivary protein expression data.
(DOC)