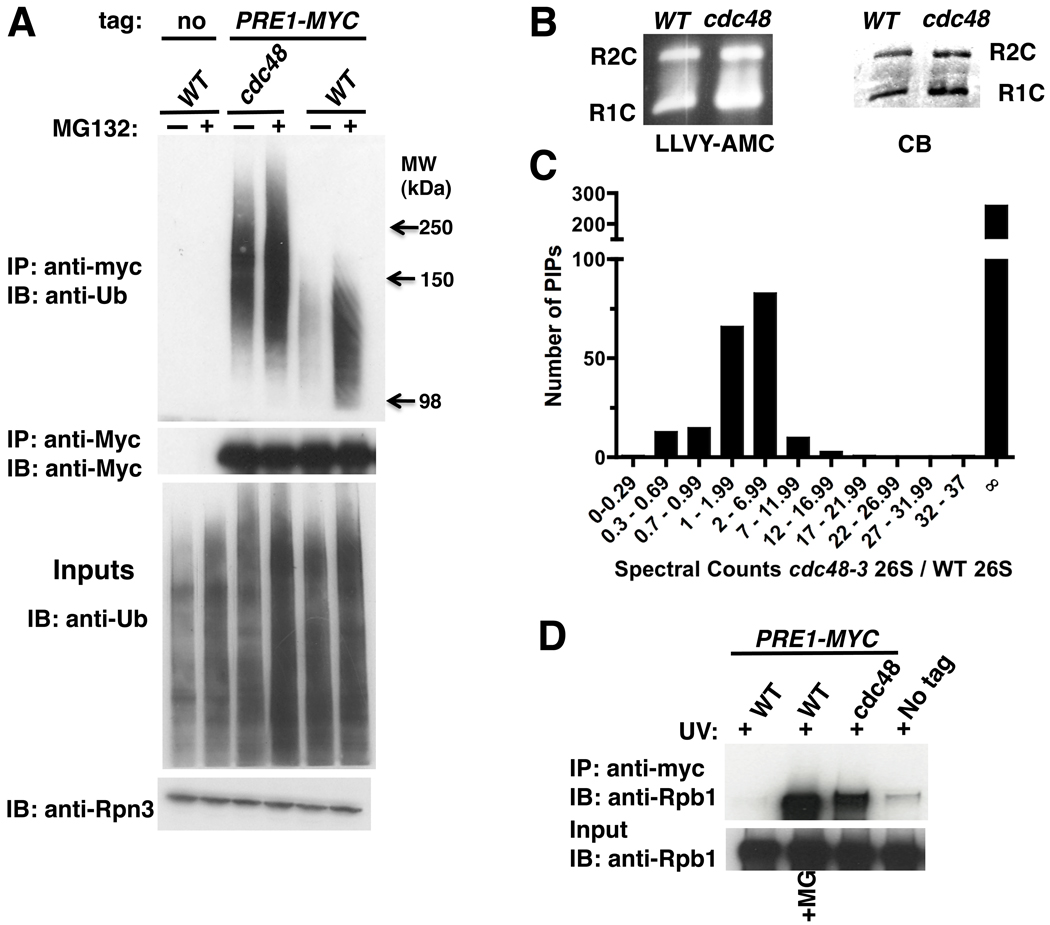
Figure 1. Ubiquitinated proteins are targeted to 26S proteasomes in cdc48-3 mutant cells.
(A) Ub conjugates accumulated on proteasomes in cdc48-3. Wildtype (WT; RJD3437) and mutant (RJD3454) cells expressing myc-tagged Pre1 (or not, RJD4090) were shifted to 37°C for 90 min and treated with 40 µM MG132 for 30 min. Proteasomes were isolated by anti-myc immunoprecipitation (IP) and immunoblotted (IB) for Ub and Pre1-myc (upper panels). Input extracts were blotted for Ub and Rpn3 (lower panels).
(B) Accumulation of conjugates on proteasome was not due to a defect in chymotryptic activity in cdc48-3 mutants. Isolated 26S proteasomes fractionated by native PAGE were either stained with Coomassie Blue (CB) or processed for in-gel peptidase activity using the fluorescent reporter substrate LLVY-AMC. R1C and R2C refer to 20S complexes capped by 1 or 2 19S regulatory particles, respectively.
(C) Numerous proteins accumulated on proteasomes in cdc48-3 mutants. Multidimensional mass spectrometry (MudPIT) was performed on an LTQ mass spectrometer using 26S proteasomes isolated from WT and mutant (RJD2902) cells grown at 37°C for 2 hours. The actual ratio of total spectral counts of proteasomal subunits from mutant to WT was 0.8 and was normalized to one. The same normalization factor was applied for spectral counts obtained for the proteasome-interacting proteins (PIPs). ∞ refers to proteins found in cdc48-3 but not WT proteasomes.
(D) Rpb1 accumulated on proteasome in cdc48-3 mutant treated with UV. WT and mutant cells were shifted to 37°C for 90 minutes and UV irradiated. Cells were recovered at 37°C for 30 minutes in the absence or presence of MG132. Input cell extracts and proteasomes immunoprecipitated (IP) with anti-myc were blotted (IB) for Rpb1 using 4H8 antibody.
See also Figure S1.