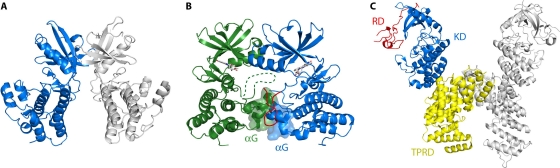
FIG. 2.
Dimerization interfaces involved in bacterial eSTK activation. (A) Back-to-back dimer revealed in the crystal structure of the M. tuberculosis PknB kinase domain (PDB accession number 1MRU). Two PknB monomers interact through a dimerization interface located in the back sides of the N-terminal lobes (130, 195). (B) Asymmetric front-to-front dimer found in the cocrystal of a mutant PknB kinase domain in complex with an ATP competitive inhibitor (PDB accession number 3F69). This structure resembles an activation complex involving the contact between the αG helixes of two monomers (103). One of the monomers (blue) shows an ordered activation loop (red), characteristic of the active state, whereas the other monomer (green) shows a disordered activation loop (represented by a dashed line). (C) Dimerization of two M. tuberculosis PknG kinase monomers. The complete structure of PknG is shown (PDB accession number 2PZI), including the N-terminal rubredoxin domain (RD) and the C-terminal tetratricopeptide repeat domain (TPRD) that surround the kinase catalytic domain (KD). In contrast with the case for PknB, dimerization of PknG occurs through interaction between the TPRDs of two monomers (153).