Abstract
The conformational structures of the hormone 17β-estradiol (E2) and the epimeric 17α-estradiol determined by solution NMR spectroscopy and restrained molecular dynamics calculations found a single low energy conformation.
Keywords: 17β-estradiol, E2, 17α-estradiol, NMR, conformation, molecular modeling
Introduction
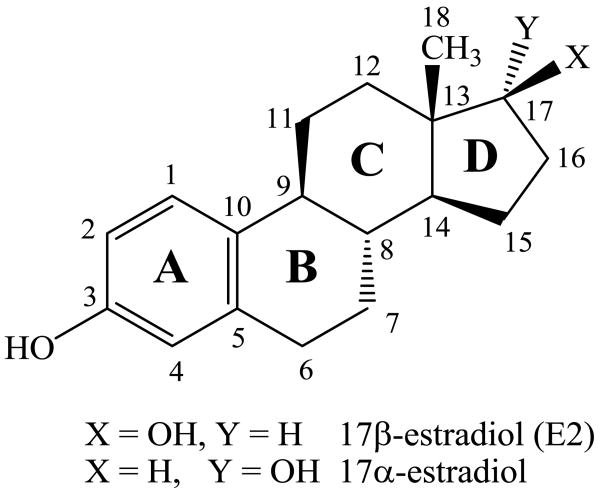
Estrogens are gonadal steroidal hormones that have important roles in reproductive function.1,2 These amphipathic steroids are quite insoluble in an aqueous environment,3,4 but circulate via the bloodstream5 signaling to a number of tissues including breast, ovaries, uterus, and brain.6 Estrogens initiate rapid nongenomic signaling events at cell membranes,2,7-10 readily diffuse across membranes interacting with nuclear estrogen receptors that regulate gene expression,6,11,12 and also act at mitochondrial membranes.13 Estrogen receptors are activated by the hormone 17β-estradiol (E2) but not by the 17α-estradiol isomer (Figure 1).14,15 Recently, the structure of the estrogen hormone 17β-estradiol (E2) as determined by NMR and X-ray was reviewed.16 In addition to the low energy C-ring chair conformation that corresponded well to crystal structures, a second conformation with a twisted boat C-ring was proposed for E2 in dimethylsulfoxide solution.16
Figure 1.
The hormone 17β-estradiol (E2) and the isomeric antiestrogen 17α-estradiol.
In conjunction with our ongoing studies of steroids interacting with phospholipid bilayer model membranes,17,18 we have also characterized the solution structures of 17β-estradiol (E2) as well as 17α-estradiol by NMR and restrained molecular dynamics calculations. Our data clearly demonstrate that for both E2 and the anomeric antiestrogen 17α-estradiol, there is only one conformation that exists in solution.
Results and discussion
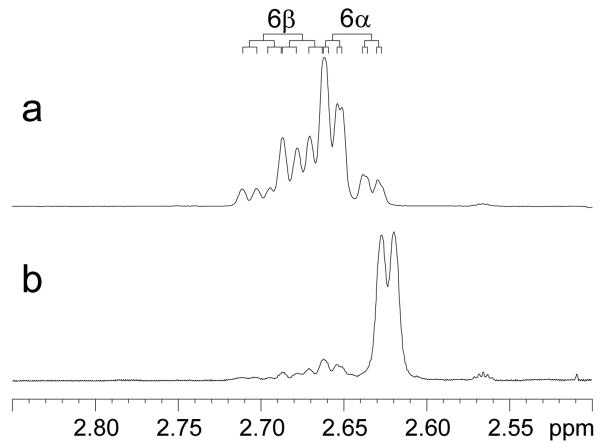
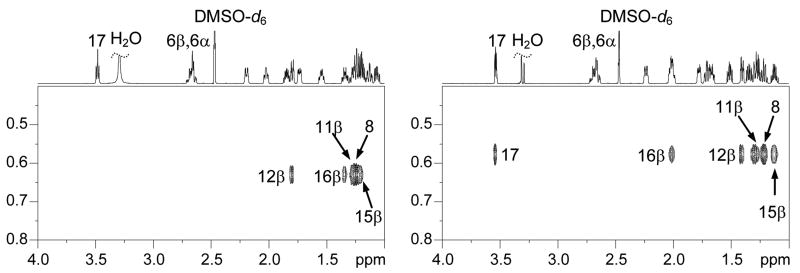
We first assigned the proton resonances of 17β-estradiol (E2) as well as 17α-estradiol by analysis of their 1D 1H-, 13C-, and 2D COSY, HSQC, and NOESY spectra recorded on a Bruker AVANCEII 700MHz NMR spectrometer (see supplementary data). All spectra were referenced to the DMSO-d6 multiplets at 2.47 ppm and 39.50 ppm. The 2D HSQC experiment was particularly useful for distinguishing resonances of H11β, H8, H15β and H7α for E2 that were between 1.19 and 1.27 ppm (see Figure 2). We have also, for the first time, distinguished the two benzylic geminal 6α and 6β protons by analyzing the J couplings of this rather complicated ABXY pattern. Our analysis was further confirmed by comparing the 1H- spectrum of E2-6β-d (Figure 3) which was synthesized in our lab according to a previously reported procedure.19
Figure 2.
1H-13C HSQC spectra of 17β-estradiol (E2, left) and 17α-estradiol (right).
Figure 3.
1H NMR of E2 (a) and E2-6β-d (b) in DMSO-d6 solution. (a) The benzylic C-6 protons of E2 appeared as an ABXY pattern with the H6β (ddd) downfield of the H6α (ddd). (b) The deuterated analog, which contained approximately 10% non-deuterated E2, appeared as a broadened doublet with a 0.03 ppm upfield shift due to the isotope effect.
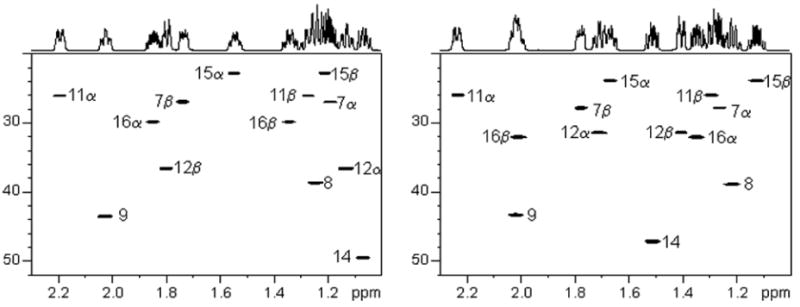
The observed proton chemical shifts, carbon chemical shifts, and the NOE intensities of key crosspeaks are tabulated for E2 and 17α-estradiol in Tables 1 and 2, respectively. Their observed J-couplings are listed in Table 3. Strong trans coplanar diaxial couplings were observed for J6β,7α, J7α,8, J8,9, J8,14, J9,11β, J11β,12α, and J14,15β. The chair conformation of the C-ring was clearly evidenced by the characteristic large J8,9, J8,14, J9,11β, and J11β,12α couplings. The C-18 methyl group gave NOEs with H8, H11β, H12β, H15β and H16β for both 17β-estradiol (E2) and 17α-estradiol, and NOE of the C-18 methyl with H17 for 17α-estradiol was also observed (Figure 4). We did not, however, observe the reported16 NOE between H12β and H15β for E2 which was due to their mis-assignment of the proton resonances. Interestingly, the NOESY of H1 with H11α in the plane of the aromatic ring gave a negative NOE, an unusal effect.20,21 This aromatic H1 proton had the expected positive NOEs with H9 and H11β, below and above the plane, analogous to H4 NOEs with H6α and H6β.
Table 1. 17β-Estradiol (E2).
| Position | Chemical Shift (ppm) | 1H-1H NOEs* | |
|---|---|---|---|
| δH | δC | ||
| 1 | 7.00 | 126.0 | 2(m); 9 (w); 11β(w) |
| 2 | 6.47 | 112.7 | 1(m) |
| 3 | — | 154.8 | |
| 4 | 6.39 | 114.9 | 6α(m); 6β(w) |
| 5 | — | 137.1 | |
| 6α | 2.65 | 29.1 | 4(m); 7α(w) |
| 6β | 2.68 | 4(w); 7β(w); 8(w) | |
| 7α | 1.19 | 26.9 | 6α(w); 7β(s); 9(m) |
| 7β | 1.74 | 6β(w); 7α(s); 8(m) | |
| 8 | 1.24 | 38.7 | 7β(m); 18(m) |
| 9 | 2.03 | 43.5 | 7α(m); 11α(w); 14(w) |
| 10 | — | 130.4 | |
| 11α | 2.19 | 26.0 | 9(w); 11β(s); 12α(w) |
| 11β | 1.27 | 11α(s); 18(m) | |
| 12α | 1.13 | 36.5 | 11α(w); 12β(s); 14(w); 17(w) |
| 12β | 1.80 | 12α(s);18(w) | |
| 13 | — | 42.8 | |
| 14 | 1.07 | 49.5 | 9(w); 12α(w); 15α(m) |
| 15α | 1.55 | 22.7 | 14(m); 15β(s); 16α(w); 16β(w) |
| 15β | 1.21 | 15α(s); 16α(w); 18(m) | |
| 16α | 1.85 | 29.8 | 15α(w); 15β(w); 16β(s); 17(m) |
| 16β | 1.34 | 15α(w); 16α(s); 17(w); 18(w) | |
| 17 | 3.48 | 80.0 | 12α(w);16α(m); 16β(w) |
| 18 | 0.63 | 11.2 | 8(m); 11β(m); 12β(w); 15β(m); 16β(w) |
NOE intensities were categorized as “strong” (s), “medium” (m) and “weak” (w), and were converted into upper limit distance constraints of 2.7, 3.5, and 5.0 Å in the molecular dynamics calculations, respectively.
Table 2. 17α-Estradiol.
| Position | Chemical Shift (ppm) | 1H-1H NOEs* | |
|---|---|---|---|
| δH | δC | ||
| 1 | 7.02 | 126.0 | 2(m); 9(w); 11β(w) |
| 2 | 6.46 | 112.6 | 1(m) |
| 3 | — | 154.8 | |
| 4 | 6.39 | 114.8 | 6α(m); 6β(w) |
| 5 | — | 137.1 | |
| 6α | 2.65 | 29.2 | 4(m); 7α(w) |
| 6β | 2.68 | 4(w); 7β(w); 8(w) | |
| 7α | 1.26 | 27.8 | 6α(w); 7β(s); 9(m) |
| 7β | 1.78 | 6β(w); 7α(s); 8(m) | |
| 8 | 1.22 | 38.8 | 7β(m); 18(m) |
| 9 | 2.02 | 43.3 | 7α(m); 11α(w); 12α(w); 14(w) |
| 10 | — | 130.4 | |
| 11α | 2.23 | 26.0 | 9(w); 11β(s); 12α(w) |
| 11β | 1.29 | 11α(s); 18(m) | |
| 12α | 1.71 | 31.4 | 9(w); 11α(w); 12β(s); 14(m) |
| 12β | 1.40 | 12α(s); 17(w); 18(w) | |
| 13 | — | 45.0 | |
| 14 | 1.51 | 47.2 | 9(w); 12α(m); 15α(m) |
| 15α | 1.67 | 23.8 | 14(m); 15β(s); 16α(w); 16β(w) |
| 15β | 1.12 | 15α(s); 16α(w); 18(w) | |
| 16α | 1.34 | 32.0 | 15α(w); 15β(w); 16β(s); 17(w) |
| 16β | 2.01 | 15α(w); 16α(s); 17(m); 18(w) | |
| 17 | 3.54 | 77.9 | 12β(w); 16α(w); 16β(m); 18(m) |
| 18 | 0.58 | 16.9 | 8(m); 11β(m); 12β(w); 15β(w); 16β(w); 17(m) |
NOE intensities were categorized as “strong” (s), “medium” (m) and “weak” (w), and were converted into upper limit distance constraints of 2.7, 3.5, and 5.0 Å in the molecular dynamics calculations, respectively.
Table 3. Observed coupling constants for 17β- and 17α-estradiol.
 |
J (Hz) |  |
J (Hz) |
|---|---|---|---|
| J1,2 | 8.5 | J1,2 | 8.4 |
| J2,4 | 2.7 | J2,4 | 2.6 |
| J6α,6β | 17.1 | J6α,6β | 17.1 |
| J6α,7α | 6.3 | J6α,7α | 6.3 |
| J6α,7β | 2.4 | J6α,7β | 2.4 |
| J6β,7α | 11.6 | J6β,7α | 11.3 |
| J6β,7β | 6.1 | J6β,7β | 6.2 |
| J7α,7β | 12.3 | J7α,7β | 12.6 |
| J7α,8 | 12.0 | J7α,8 | 12.0 |
| J7β,8 | 2.3 | J7β,8 | 2.5 |
| J8,9 | 11.2 | J8,9 | 11.2 |
| J8,14 | 12.4 | J8,14 | 12.2 |
| J9,11α | 4.3 | J9,11α | 4.3 |
| J9,11β | 11.2 | J9,11β | 11.0 |
| J11α,11β | 13.5 | J11α,11β | 13.4 |
| J11α,12α | 4.1 | J11α,12α | 4.3 |
| J11α,12β | 2.9 | J11α,12β | 2.7 |
| J11β,12α | 12.8 | J11β,12α | 13.2 |
| J11β,12β | 3.8 | J11β,12β | 4.0 |
| J12α,12β | 12.7 | J12α,12β | 13.0 |
| J14,15α | 7.4 | J14,15α | 7.4 |
| J14,15β | 10.8 | J14,15β | 10.8 |
| J15α,15β | 12.1 | J15α,15β | 12.1 |
| J15α,16α | 9.4 | J15α,16α | 9.5 |
| J15α,16β | 3.4 | J15α,16β | 2.9 |
| J15β,16α | 5.8 | J15β,16α | 6.6 |
| J15β,16β | 11.5 | J15β,16β | 12.2 |
| J16α,16β | 13.4 | J16α,16β | 14.3 |
| J16α,17α | 8.9 | J16α,17β | 0.0 |
| J16β,17α | 8.4 | J16β,17β | 5.8 |
Figure 4.
NOESY spectra of 17β-estradiol (E2, left) and 17α-estradiol (right).
Our NOE data in Tables 1 and 2 were used as distance restraints to determine the conformations of 17β-estradiol (E2) and 17α-estradiol, respectively. The calculations were performed using Macromodel in the Schrodinger software package. In order to fully sample the entire conformational space, stochastic dynamics simulations with NOE distance restraints were carried out at 1000K with a time step of 1.0 fs. Conformations were recorded every 250,000 steps for a total of 20 trajectories. Each conformer was further subject to conjugate gradient minimization until the maximum derivative was less than 0.05 kJ/mol. Figure 5a depicts the 20 conformers of 17β-estradiol (E2) and Figure 5b depicts the 20 conformers of 17α-estradiol. For each compound, all 20 conformers were closely superimposable over the entire backbone. The solution conformations of the anomeric steroids were in good agreement with reported X-ray structures22-26 and included the following features. The aromatic A-ring was planar. The unsaturated B-ring had trans coplanar diaxial orientations for H6β with H7α and for H8 with H9 and was a half-chair (Figure 5c). The C-ring was a chair conformation with trans coplanar diaxial orientations for H11β with H9 and H12α as well as for H8 with H9 and H14 (Figure 5d). Thus, the conformations of the ABC rings of both anomers were nearly identical. The five-membered D-rings were both C-13 β-envelopes with only 3° of distortion (C18-C13-C17 was 110.3° for E2 and 107.3° for the α-anomer, Figures 5e and 5f, respectively) due to the steric interaction between the C-18 methyl group and the C-17 hydroxyl group of E2.
Figure 5.
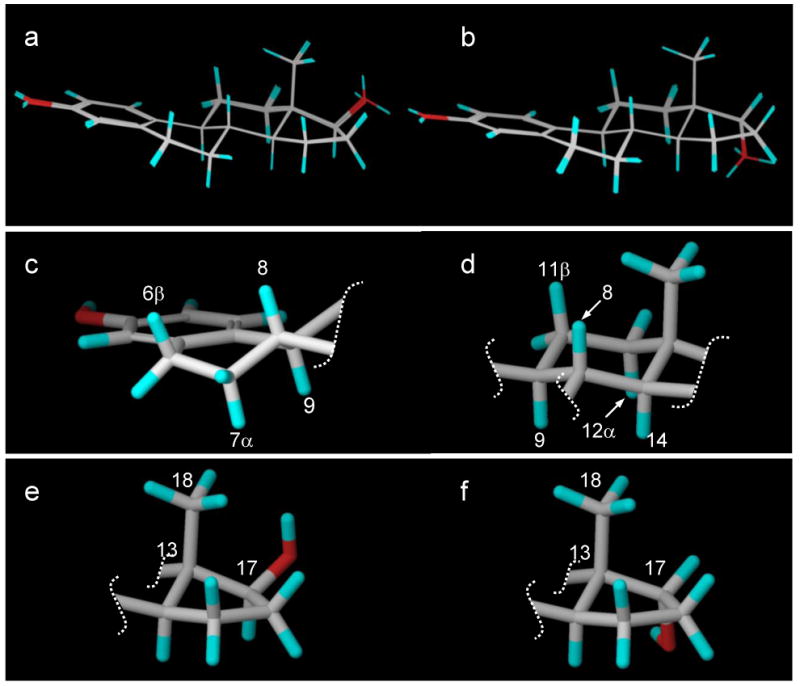
(a) The 20 low-energy conformers of 17β-estradiol (E2). (b) The 20 low-energy conformers of 17α-estradiol. (c) The unsaturated B-ring half-chair showing trans coplanar diaxial orientations for H6β with H7α and for H8 with H9. (d) The C-ring chair with trans coplanar diaxial orientations for H11β with H9 and H12α as well as for H8 with H9 and H14. (e) The D-ring for 17β-estradiol (E2). (f) The D-ring for 17α-estradiol.
The conformational structures of the hormone 17β-estradiol (E2) and the anomeric antiestrogen 17α-estradiol have now been clearly established in DMSO solution using a combination of NMR spectroscopy and restrained molecular dynamics calculations. We have found that there is only one low energy conformation for theses steroids with a chair conformation for their C-rings. We have also, for the first time, assigned all resonances including the two benzylic geminal H6α and H6β protons. We are currently using these structural details to study the interactions of these compounds with bicelle model membranes.
Supplementary Material
Acknowledgments
This work was supported by grants DA-152, DA-3801, DA-7215, DA-9158, and DA-24842 from the National Institute on Drug Abuse.
Abbreviations
- COSY
correlation spectroscopy
- E2
17β-estradiol
- HSQC
heteronuclear single quantum coherence
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser enhancement spectroscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carrer HF, Cambiasso MJ. Cell Mol Neurobiol. 2002;22:479. doi: 10.1023/A:1021825317546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Micevych P, Kuo J, Christensen A. J Neuroendocrinol. 2009;21:249. doi: 10.1111/j.1365-2826.2009.01833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurwitz AR, Liu ST. J Pharm Sci. 1977;66:624. doi: 10.1002/jps.2600660504. [DOI] [PubMed] [Google Scholar]
- 4.Ilardia-Arana D, Kristensen HG, Mullertz A. J Pharm Sci. 2006;95:248. doi: 10.1002/jps.20494. [DOI] [PubMed] [Google Scholar]
- 5.Jacobsohn GM, Siegel ET, Jacobsohn MK. J Clin Endocrinol Metab. 1975;40:177. doi: 10.1210/jcem-40-2-177. [DOI] [PubMed] [Google Scholar]
- 6.Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M, Parker MG, Gustafsson JA. Pharmacol Rev. 2006;58:773. doi: 10.1124/pr.58.4.8. [DOI] [PubMed] [Google Scholar]
- 7.Hammes SR, Levin ER. Endo Rev. 2007;28:726. doi: 10.1210/er.2007-0022. [DOI] [PubMed] [Google Scholar]
- 8.Prossnitz ER, Maggiolini M. Mol Cell Endocrinol. 2009;308:32. doi: 10.1016/j.mce.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olde B, Leeb-Lundberg LMF. Trends Endocrinol Metab. 2009;20:409. doi: 10.1016/j.tem.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Boulware MI, Mermelstein PG. Steroids. 2009;74:608. doi: 10.1016/j.steroids.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Physiol Rev. 2001;81:1535. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 12.Ellmann S, Sticht H, Thiel F, Beckmann MW, Strick R, Strissel PL. Cell Mol Life Sci. 2009;66:2405. doi: 10.1007/s00018-009-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simpkins JW, Dykens JA. Brain Res Rev. 2008;57:421. doi: 10.1016/j.brainresrev.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Wiese TE, Polin LA, Palomino E, Brooks SC. J Med Chem. 1997;40:3659. doi: 10.1021/jm9703294. [DOI] [PubMed] [Google Scholar]
- 15.Toran-Allerand CD, Tinnikov AA, Singh RJ, Nethrapalli IS. Endocrinology. 2005;146:3843. doi: 10.1210/en.2004-1616. [DOI] [PubMed] [Google Scholar]
- 16.Commodari F, Sclavos G, Ibrahimi S, Khiat A, Boulanger Y. Magn Reson Chem. 2005;43:444. doi: 10.1002/mrc.1581. [DOI] [PubMed] [Google Scholar]
- 17.Makriyannis A, Fesik S. J Med Chem. 1983;26:463. doi: 10.1021/jm00358a001. [DOI] [PubMed] [Google Scholar]
- 18.Mavromoustakos T, Yang DP, Makriyannis A. Biochim Biophys Acta. 1994;1194:69. doi: 10.1016/0005-2736(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 19.Debrauwer L, Rathahao E, Jouanin I, Paris A, Clodic G, Molines H, Convert O, Fournier F, Tabet JC. J Am Soc Mass Spectrom. 2003;14:364. doi: 10.1016/S1044-0305(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 20.Mersh JD, Sanders JKM. Org Magn Reson. 1982;18:122. [Google Scholar]
- 21.Szántay C., Jr Bull Magn Reson. 1992;14:112. [Google Scholar]
- 22.Busetta B, Hospital M. C R Acad Sci Paris, Ser C. 1969;268:1300. [Google Scholar]
- 23.Busetta B, Barrans Y, Precigoux G, Hospital M. Act Cryst B. 1976;32:1290. [Google Scholar]
- 24.Palomino E, Heeg MJ, Horwitz JP, Brooks SC. J Steroid Biochem. 1990;35:219. doi: 10.1016/0022-4731(90)90278-z. [DOI] [PubMed] [Google Scholar]
- 25.Wiese TE, Brooks SC. J Steroid Biochem Mol Biol. 1994;50:61. doi: 10.1016/0960-0760(94)90173-2. [DOI] [PubMed] [Google Scholar]
- 26.Lamminmaki U, Kankare JA. J Biol Chem. 2001;276:36687. doi: 10.1074/jbc.M102367200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.