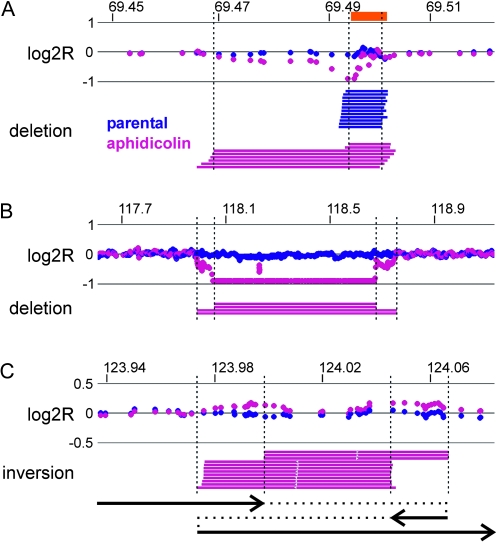
Figure 1.—
Examples of aphidicolin-induced CNVs. Intensity data (log2R) from SNP arrays are at the top of each panel with mate pairs of the indicated type below. Array data are averaged over a five-probe window. Individual mate pairs are drawn as bars connecting the two reads as mapped to the reference genome. Blue denotes the reference 090 sample and magenta the aphidicolin-treated A3A2 sample. Chromosome coordinates are in megabases. (A) An induced deletion on chromosome 15 that overlaps a constitutional CNV. The orange box denotes the position of a known human population CNV. (B) A bi-allelic deletion on chromosome 3. (C) A complex inversion on chromosome 4 that creates two associated copy-number gains. One potential underlying allele is drawn as an event chain similar to Figure S7.