Abstract
Phenotypic switching has been observed in laboratory studies of yeast and bacteria, in which the rate of such switching appears to adjust to match the frequency of environmental changes. Among possible mechanisms of switching are epigenetic influences on gene expression and variation in levels of methylation; thus environmental and/or genetic factors may contribute to the rate of switching. Most previous analyses of the evolution of phenotypic switching have compared exponential growth rates of noninteracting populations, and recombination has been ignored. Our genetic model of the evolution of switching rates is framed in terms of a mutation-modifying gene, environments that cause periodic changes in fitness, and recombination between the mutation modifier and the gene under selection. Exact results are obtained for all recombination rates and symmetric fitnesses that strongly generalize earlier results obtained under complete linkage and strong constraints on the relation between fitness and period of switching. Our analytical and numerical results suggest a general principle that recombination reduces the stable rate of switching in symmetric and asymmetric fitness regimes and when the period of switching is random. As the recombination rate increases, it becomes less likely that there is a stable nonzero rate of switching.
IN large populations subject to mutation between alleles under selection that is constant over time, mutation–selection equilibria can be stable. In the neighborhood of such a stable equilibrium, if an allele at a locus that controls the rate of mutation is introduced, this allele will invade if it reduces the mutation rate (Liberman and Feldman 1986). This is one example of the reduction principle for constant selection regimes (Feldman and Liberman 1986). This approach, using the selectively neutral genetic modifiers of parameters such as mutation, recombination, and migration that are important features of the evolutionary process, can be viewed as an alternative to an approach using evolutionary stable strategies (ESS) (Maynard Smith 1978) or to finding critical points of the mean fitness (Karlin and McGregor 1972, 1974).
It has become usual to denote the gene on which selection occurs directly as the major gene (or in the case of recombination modification, genes) and the locus that controls the parameter of interest as the modifier gene (e.g., Feldman et al. 1997). If a new mutation at the modifier gene is introduced during a transient phase of evolution, rather than near an equilibrium, the fate of the modifier mutant may be quite different: for example, an allele that causes reduction of recombination will succeed if it is introduced near stable linkage disequilibrium, but if it arises at a phase of the dynamics where the major loci are proceeding toward fixation, increase of recombination may occur and the reduction principle does not necessarily hold (Maynard Smith 1980, 1988; Bergman and Feldman 1990). The evolution of modifiers of mutation, recombination, or migration rates when the regime of selection on the major gene(s) is not constant over time has seen far less mathematical or even numerical analysis than the case of constant selection. Early numerical work by Charlesworth (1976) showed the failure of the reduction principle for recombination under some patterns of cyclically fluctuating abiotic selection.
Host–parasite systems may produce cyclical dynamics that have features similar to those of cyclically fluctuating abiotic selection. The evolution of host recombination in a host–parasite cyclical system has been addressed by Hamilton (1980) and most recently by Gandon and Otto (2007). Hamilton used a mean fitness argument to demonstrate an advantage to hosts with higher recombination, and Nee (1989) used a similar approach in finding that mutation rates should increase in both host and parasite. Gandon and Otto (2007) showed that alleles at a recombination-modifying locus that increased recombination could succeed in such host–parasite models. Mutation-increasing alleles may be favored in similar models, as shown by Haraguchi and Sasaki (1996) and M'Gonigle et al. (2009). The latter authors also showed that under host–parasite cycling, increasing recombination between the mutation modifier and the gene under selection in the host decreased the stable mutation rate.
With increasing empirical interest in epigenetics, there has been a flurry of activity surrounding “stochastic switching,” a phenomenon observed in some studies of Saccharomyces cerevisiae, Escherichia coli, or Bacillus subtilis, where individual cells switch cyclically between different inheritable phenotypes (Thattai and van Oudenaarden 2004; Kussell and Leibler 2005; Acar et al. 2008). For example, experiments by Balaban et al. (2004) show that switching between phenotypes may differ between different genetic strains of E. coli. The same group analyzed a mathematical model of this system and showed that the optimal rate of phenotypic switching was strongly dependent on the frequency of environmental changes but only weakly dependent on the strength of selection in any single environment (Kussell et al. 2005). Earlier theoretical treatments by Ishii et al. (1989) and Lachmann and Jablonka (1996) also found that optimal switching rates depended on the distribution of environmental periodicities.
Phenotypic switching may be an example of an epigenetic effect, for example based on methylation (Lim and van Oudenaarden 2007). There is some discussion as to the fraction of such epigenetic effects that are transgenerational (e.g., Youngson and Whitelaw 2008) and, to the extent that they are, what their evolutionary effects might be (Bonduriansky and Day 2009). Most theoretical treatments of phenotypic switching have assumed that the organism is asexual and that the numbers of individuals grow exponentially (e.g., Kussell et al. 2005, p. 1809; Gaál et al. 2010). These studies usually do not specifically include genetic contributions to the rate of switching; exceptions are the analyses by Ishii et al. (1989) and Salathé et al. (2009), both of which took fitness to be the phenotype that switches and allowed the rate of switching to be under the genetic control of a mutation-modifying locus. If switching is epigenetic and heritable, it is reasonable to propose that it is at least partially under genetic control. Further, if phenotypic switching involves epigenetic regulation of gene expression, and if this epigenetic phenomenon occurs in sexual species (e.g., Rakyan et al. 2003; Henderson and Jacobsen 2007), it is also reasonable to investigate the importance of recombination between genes contributing to the phenotype and those contributing to the rate of switching. As pointed out by Lynch (2007, p. 89, Table 4.1) the importance of recombination in the evolution of prokaryotes has been substantially underestimated. Inclusion of the mutation modifier and recombination carries the evolution of phenotypic switching into the corpus of evolutionary population genetics.
One of the earliest studies of this problem in the framework of theoretical population genetics was by Ishii et al. (1989), who studied a haploid locus with two alleles A and a, whose fitnesses were 1 + s(t) and 1 − s(t), respectively, at generation t with s(t) allowed to fluctuate through time with average zero. A second locus with alleles B and b controlled the (bidirectional) mutation rate between A and a, and the recombination rate between the two genes was r. Selection changed cyclically. In the case of complete linkage (r = 0), they found that the ESS mutation rate maximized the long-term geometric mean population fitness. For r > 0 their analysis depended on the symmetric selection coefficient, s, being ≪1/n, where 2n is the period of the selection cycle. The recombination r also appeared through the size of the product rn. Their numerical analysis suggested that in this symmetric case, no matter what the value of r, if selection was strong enough (s large enough), the value 1/n for the mutation rate could not be invaded. The analysis by Lachmann and Jablonka (1996) did not involve a modifier (and hence recombination was irrelevant) but found, similarly to Ishii et al. (1989), that in the symmetric selection case the (fitness) optimum mutation rate was ∼1/n. They also claimed that this result held in asymmetric fitness regimes. An important qualitative interpretation of this result is that if the mutation rate is initially low enough, a modifier allele that increases the mutation rate can invade. Thus, violations of the Feldman–Liberman reduction principle (Feldman and Liberman 1986) for constant environments are to be expected in cyclically varying environments.
A numerical treatment by Salathé et al. (2009) of the symmetric case confirmed the evolutionary stability of a mutation rate of ∼1/n in a deterministically cycling environment of period n generations. However, if the time in each environment is random, then even in the symmetric case the stable mutation rate can be vastly different from 1/n (Salathé et al. 2009, Figure 1). Further, if the fitnesses of A relative to a in environment 1 and a relative to A in environment 2 are not exactly the same, i.e., there is no symmetry, then for a wide range of selection coefficients and initial mutation rates, a mutation-reducing modifier allele will succeed, potentially taking the switching rate to zero. Gaál et al. (2010) proved a parallel set of results in the asexual exponential growth framework.
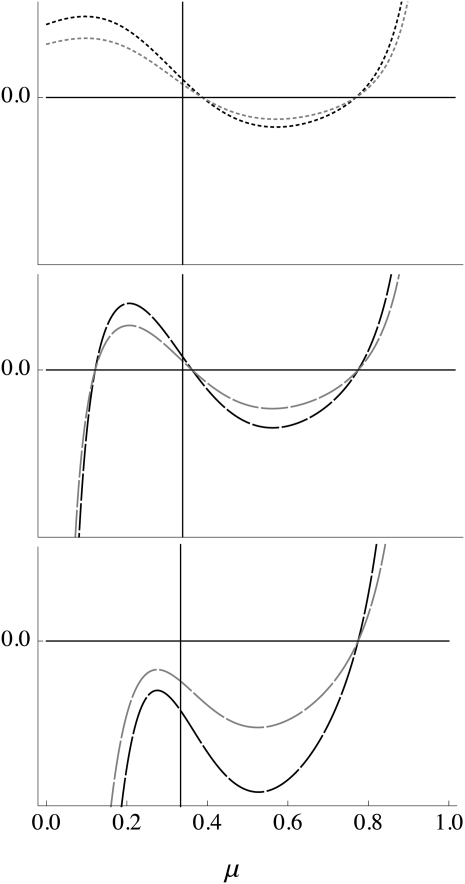
Figure 1.—
Comparison of 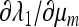 evaluated at μm = μM = μ (solid curves) and
evaluated at μm = μM = μ (solid curves) and 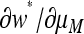 (shaded curves) for n = 3. In all plots, s1 = 10−1 whereas s3 = 10−1, 10−0.8, and 10−0.6, in the top, middle, and bottom plots, respectively. The vertical line is located at μ = 1/n =
(shaded curves) for n = 3. In all plots, s1 = 10−1 whereas s3 = 10−1, 10−0.8, and 10−0.6, in the top, middle, and bottom plots, respectively. The vertical line is located at μ = 1/n = 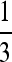 .
.
Our goal in this article is to study mutation modification as a mechanism for the control of phenotypic switching where the phenotype is fitness. We obtain complete analytic results for period 2n = 2 or 2n = 4 with symmetric fitnesses and recombination, where the stable values of the mutation rate μ are 1.0 and 0.5, respectively. We also investigate an interesting property of the period, namely that there is a mathematical difference between the cases in which the selection regime changes after an odd or an even number of generations. We find that in general the presence of recombination makes it more unlikely that there is a stable nonzero mutation rate, whether the cycle period is fixed or random. As the rate of recombination increases, the extent of departure from symmetric fitnesses that permits a stable nonzero mutation rate becomes smaller.
REFERENCE MODEL: CONSTANT ENVIRONMENT
Consider a population of haploids large enough that genetic drift can be ignored. Fitness is determined by the alleles A and a at the major locus, and linked to this locus, with recombination fraction r, is a modifier locus with alleles M and m that produce mutation rates μM and μm, respectively. The mutation rates from A to a and a to A are the same. The four genotypes AM, Am, aM, and am have frequencies x1, x2, x3, and x4, respectively, and fitnesses w1, w2, w3, and w4, where, because the modifier locus is selectively neutral, we have w1 = w2 and w3 = w4. Then the frequencies 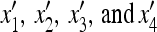 in the next generation are
in the next generation are
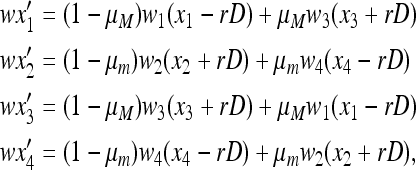 |
(1) |
where D = x1x4 − x2x3 is the linkage disequilibrium, and the normalizing factor w is
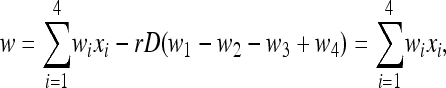 |
(2) |
because of our fitness assumptions. We have assumed that the life cycle begins with a diploid phase during which there is Mendelian segregation with recombination followed by selection on the haploid phase after which there is mutation. Recombination, or homologous exchange, is an important step in this life cycle for a vast array of organisms, including microbes (Lynch 2007, pp. 88–95).
In the absence of the modifier allele m (in which case recombination has no effect), the transformation (1) is linear fractional; that is, the transformation is of the form 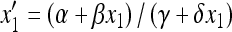 , where α, β, γ, and δ are constants, and we have used x3 = (1 − x1). This allows complete determination of the global dynamics of (x1, 0, x3, 0). We can summarize these dynamics as follows. When allele M is fixed, there is a unique globally stable equilibrium
, where α, β, γ, and δ are constants, and we have used x3 = (1 − x1). This allows complete determination of the global dynamics of (x1, 0, x3, 0). We can summarize these dynamics as follows. When allele M is fixed, there is a unique globally stable equilibrium 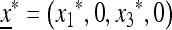 , where
, where 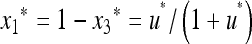 and u* is the unique positive root of
and u* is the unique positive root of
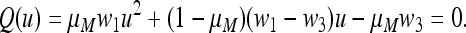 |
(3) |
We have 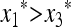 if (1 − 2μM)(w1 − w3) > 0 and
if (1 − 2μM)(w1 − w3) > 0 and 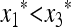 if (1 − 2μM)(w1 − w3) < 0.
if (1 − 2μM)(w1 − w3) < 0.
The evolution of mutation is determined by the stability of 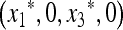 to the introduction of allele m, that is, whether m will increase in frequency when introduced close to this equilibrium. The local stability of this equilibrium to invasion by Am and am is determined by the linear transformation
to the introduction of allele m, that is, whether m will increase in frequency when introduced close to this equilibrium. The local stability of this equilibrium to invasion by Am and am is determined by the linear transformation
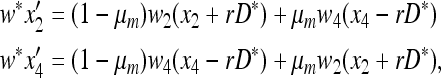 |
(4) |
where
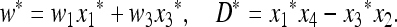 |
(5) |
From (4), the matrix 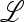 associated with this linear transformation is
associated with this linear transformation is
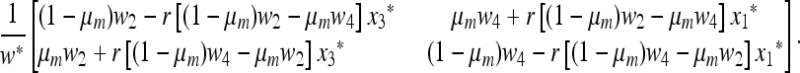 |
(6) |
The stability of this point is determined by the eigenvalues of 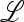 , which satisfy the equation
, which satisfy the equation 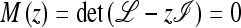 , where
, where 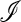 is the 2 × 2 identity matrix. The positive eigenvalue of
is the 2 × 2 identity matrix. The positive eigenvalue of 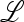 is <1 if M(1) > 0 and M′(1) > 0 and is >1 if M(1) < 0. In the former case, m cannot invade, while in the latter case
is <1 if M(1) > 0 and M′(1) > 0 and is >1 if M(1) < 0. In the former case, m cannot invade, while in the latter case 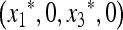 is unstable and m invades. After a considerable amount of algebra we find the following.
is unstable and m invades. After a considerable amount of algebra we find the following.
Result 1. The equilibrium 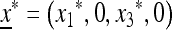 is stable if μm > μM and unstable if μm < μM. For all recombination rates, a modifier allele that reduces the mutation rate will invade.
is stable if μm > μM and unstable if μm < μM. For all recombination rates, a modifier allele that reduces the mutation rate will invade.
We can also show that the mean fitness w* at 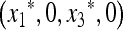 is a decreasing function of μM.
is a decreasing function of μM.
This reduction principle in constant environments was proved for selection on diploids and any number of alleles at the mutation-controlling gene by Liberman and Feldman (1986).
CHANGING ENVIRONMENTS: INITIAL EQUILIBRIUM
Assume that in each generation the genotypic fitnesses change. At generation i, for i = 1, 2,…, k, the fitness parameters are
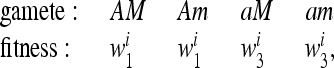 |
(7) |
where again we assume M and m do not affect fitness.
At the beginning of generation i, for i = 1, 2,…, k, the population state is given by the frequency vector 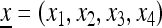 , and at the beginning of generation i + 1 it is given by
, and at the beginning of generation i + 1 it is given by 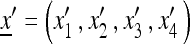 , where
, where 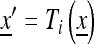 and Ti is given by the system (1) with fitnesses (7):
and Ti is given by the system (1) with fitnesses (7):
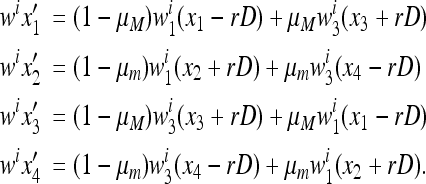 |
(8) |
The normalizing factor is 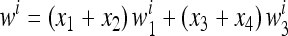 and the linkage disequilibrium is
and the linkage disequilibrium is 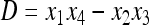 .
.
If the population state at the beginning of the k-generation process is 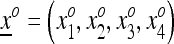 , then after generation k it is
, then after generation k it is 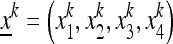 where
where 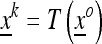 and the transformation T is given by the composition of the k transformations:
and the transformation T is given by the composition of the k transformations:
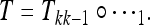 |
(9) |
The combined mean fitness associated with the transformation T from 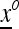 to
to 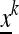 is
is
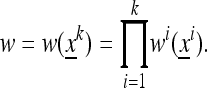 |
(10) |
To study the evolution of mutation, we first consider a population where only the M allele is present at the modifier locus. Let Si be the transformation associated with Ti restricted to the two gametes AM and aM only. Then Si, for i = 1, 2,…, k, is given by
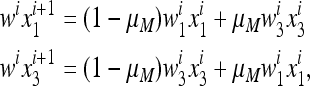 |
(11) |
with wi given by
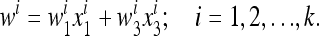 |
(12) |
The “total” k-generation transformation restricted to AM and aM is 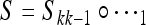 .
.
In (11) we can use the ratio 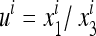 instead of
instead of 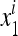 and
and 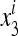 , and similarly
, and similarly 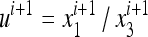 . Then the transformation Si is
. Then the transformation Si is
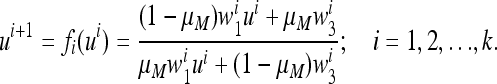 |
(13) |
Thus, the total k-generation transformation on the boundary where only M is present is the composition 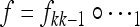 .
.
Note that 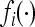 is a linear fractional transformation. One of the properties of such a transformation is that its composition over k generations is also a linear fractional transformation. Hence the total k-generation transformation f can be written as
is a linear fractional transformation. One of the properties of such a transformation is that its composition over k generations is also a linear fractional transformation. Hence the total k-generation transformation f can be written as
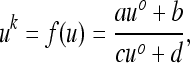 |
(14) |
where, since the coefficients in (13) are positive for 0 < μM < 1, the coefficients a, b, c, and d are also positive. Now we can analyze the evolutionary process in terms of this k-generation transformation (14), and since it is linear fractional it converges to a unique stable equilibrium. Thus, ultimately the population converges to a k-step trajectory loop determined by the stable state of this k-step compound linear fractional transformation. In fact, u*, the stable state of the k-step process, is the unique positive root of
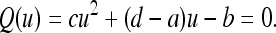 |
(15) |
Using the same notation as in Result 1, this equilibrium can be expressed as 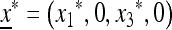 , and it can be shown to be globally stable.
, and it can be shown to be globally stable.
CHANGING ENVIRONMENTS: EXTERNAL STABILITY
To explore the dynamics of switching, and in particular to find an evolutionarily stable switching rate (if it exists), we study the external stability of the unique internally stable equilibrium 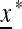 to invasion by the new mutation modifying allele m. The mutation rate μM produced by allele M will be evolutionarily stable if allele m cannot invade when μm > μM or μm < μM. Therefore we analyze the external stability of
to invasion by the new mutation modifying allele m. The mutation rate μM produced by allele M will be evolutionarily stable if allele m cannot invade when μm > μM or μm < μM. Therefore we analyze the external stability of 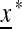 as a function of the difference between the new mutation rate μm and the resident rate μM. This will give us the direction of evolution of the switching.
as a function of the difference between the new mutation rate μm and the resident rate μM. This will give us the direction of evolution of the switching.
A population starting from the equilibrium 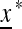 satisfies
satisfies 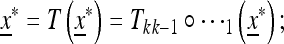 that is, after k generations of change it will end up again at x*. Take
that is, after k generations of change it will end up again at x*. Take 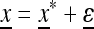 as a “starting” population state near
as a “starting” population state near 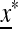 , where
, where 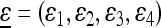 with ɛi “small” and
with ɛi “small” and 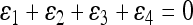 so that
so that 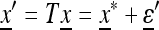 . We work with the linear transformation “near”
. We work with the linear transformation “near” 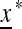 ; that is, up to nonlinear terms,
; that is, up to nonlinear terms,
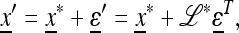 |
(16) |
where 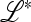 is a matrix that depends on
is a matrix that depends on 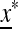 and may be obtained as a combination of the matrices of similar linear approximations across the k generations. The stability of equilibrium
and may be obtained as a combination of the matrices of similar linear approximations across the k generations. The stability of equilibrium 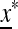 to the introduction of allele m is determined by the eigenvalues of the matrix
to the introduction of allele m is determined by the eigenvalues of the matrix 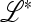 .
.
This scenario is well known from modifier theory (Feldman 1972; Feldman et al. 1980; Feldman and Liberman 1986), and the matrix 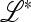 is also known to have the structure
is also known to have the structure
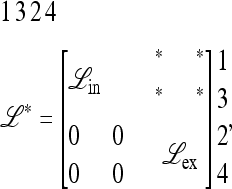 |
(17) |
where we have swapped columns 2 and 3 and rows 2 and 3 to show the structure. The entries marked * do not affect the eigenvalues of 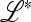 .
.
The eigenvalues of 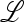 are therefore those of the submatrices
are therefore those of the submatrices 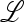 in and
in and 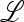 ex, where
ex, where 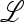 in determines the internal stability of
in determines the internal stability of 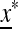 , confined to the boundary with only M present. As
, confined to the boundary with only M present. As 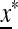 is assumed to be stable there, these eigenvalues are less than one in magnitude.
is assumed to be stable there, these eigenvalues are less than one in magnitude.  ex is the linear approximation of evolution near
ex is the linear approximation of evolution near 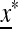 , which involves only the gametes Am and am and is a combination of the matrices
, which involves only the gametes Am and am and is a combination of the matrices 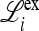 , for i = 1, 2,…, k, that describe the changes in generation i when allele m is rare.
, for i = 1, 2,…, k, that describe the changes in generation i when allele m is rare.
In fact, from (8) we have
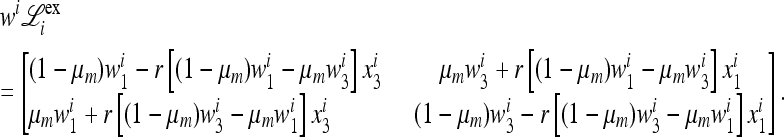 |
(18) |
We set 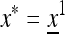 and describe the following generations by
and describe the following generations by 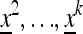 . The normalizer in generation i is
. The normalizer in generation i is
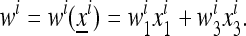 |
(19) |
As each of the matrices 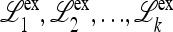 is positive,
is positive,  ex is also positive, and, by the Perron–Frobenius theory, the largest eigenvalue of
ex is also positive, and, by the Perron–Frobenius theory, the largest eigenvalue of  ex is positive. Some properties of
ex is positive. Some properties of 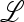 are documented in supporting information, File S1, Part I.
are documented in supporting information, File S1, Part I.
The external stability of 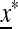 depends on the magnitude of the positive, and largest, so-called Perron–Frobenius eigenvalue of
depends on the magnitude of the positive, and largest, so-called Perron–Frobenius eigenvalue of 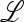 ex. If this eigenvalue is <1,
ex. If this eigenvalue is <1, 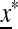 is externally stable and allele m cannot invade near
is externally stable and allele m cannot invade near 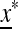 , and if it is >1,
, and if it is >1, 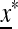 is externally unstable, and m enters the population and increases in frequency.
is externally unstable, and m enters the population and increases in frequency.
The characteristic polynomial of 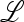 ex, namely C(z) = det(
ex, namely C(z) = det(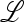 ex − z
ex − z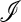 ), is quadratic in z with a positive z2 coefficient. Therefore the largest positive eigenvalue of
), is quadratic in z with a positive z2 coefficient. Therefore the largest positive eigenvalue of 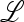 ex is <1 when C(1) > 0 and C′(1) > 0, and it is >1 when C(1) < 0. It seems impossible to compute C(1) and C′(1) in general. We are, however, able to derive analytical results in some special cases that are discussed in the following sections.
ex is <1 when C(1) > 0 and C′(1) > 0, and it is >1 when C(1) < 0. It seems impossible to compute C(1) and C′(1) in general. We are, however, able to derive analytical results in some special cases that are discussed in the following sections.
CYCLICALLY FLUCTUATING ENVIRONMENTS: PERIOD 2
Suppose that the selection regime alternates between two states as specified in (20) below.
 |
(20) |
Environments alternate, producing a cycle with period 2.
We first characterize the unique stable equilibrium 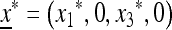 on the boundary where only M is present. On this boundary the two transformations S1 and S2 of (11) are
on the boundary where only M is present. On this boundary the two transformations S1 and S2 of (11) are
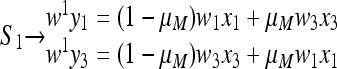 |
(21) |
and
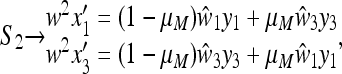 |
(22) |
where x1 and x3 are the frequencies of AM and aM, respectively, at the beginning of generation 1, y1 and y3 at the end of generation 1, and 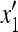 and
and 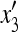 at the end of generation 2. The mean fitnesses are
at the end of generation 2. The mean fitnesses are
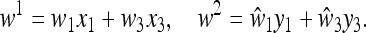 |
(23) |
This cycle repeats with period 2, and the combined one-cycle transformation 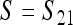 , on the boundary where M is fixed, is
, on the boundary where M is fixed, is
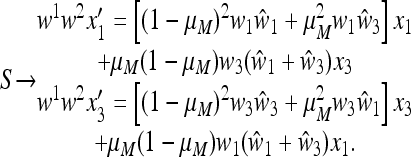 |
(24) |
The normalizing factor w = w1w2 is the cycle mean fitness and can be expanded as
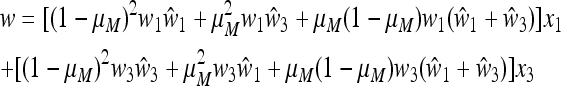 |
(25) |
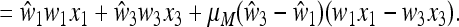 |
(26) |
The linear fractional transformation during the two generations, S, described in (14), is
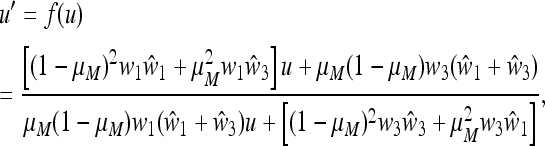 |
(27) |
where 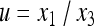 and
and 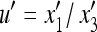 .
.
The stable equilibrium 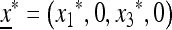 with
with 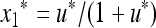 and
and 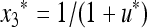 satisfies u* = u′ = f(u*) and is the unique positive root of the quadratic equation Q(u) = 0, where
satisfies u* = u′ = f(u*) and is the unique positive root of the quadratic equation Q(u) = 0, where
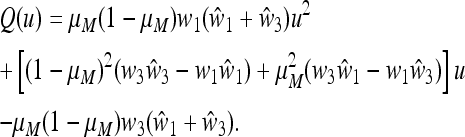 |
(28) |
We now check the external stability properties of 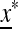 , that is, its stability to invasion by m when m is introduced near x*. Evolution of the mutation rate is determined by the external stability properties of
, that is, its stability to invasion by m when m is introduced near x*. Evolution of the mutation rate is determined by the external stability properties of 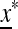 , that is, its stability to invasion by m when m is introduced near
, that is, its stability to invasion by m when m is introduced near 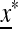 . In File S1 we show that m will invade (i.e., μm invades μM) if M(1) < 0, where
. In File S1 we show that m will invade (i.e., μm invades μM) if M(1) < 0, where
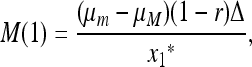 |
(29) |
and Δ = Δ(r, μm) is shown in File S2 to be a bilinear function of r and μm in 0 ≤ r ≤ 1 and 0 ≤ μm ≤ 1.
In general it is very complicated to compute M(1), unless we assume that the fitnesses are symmetric; that is, 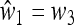 and
and 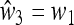 , which allows a complete analysis of M(1) and the external stability of
, which allows a complete analysis of M(1) and the external stability of 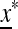 . For general r, μm, and μM, we refer to Numerical Analysis, below. We are able to obtain complete results when there is absolute linkage between the two loci (r = 0) and μm = 0 or μm = 1. When r = 0, the matrix
. For general r, μm, and μM, we refer to Numerical Analysis, below. We are able to obtain complete results when there is absolute linkage between the two loci (r = 0) and μm = 0 or μm = 1. When r = 0, the matrix 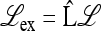 , with w* the product of w1 and w2 at equilibrium, is given in File S1, Part II by
, with w* the product of w1 and w2 at equilibrium, is given in File S1, Part II by
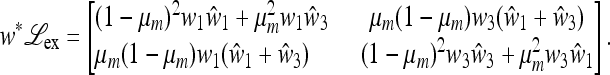 |
(30) |
When either μm = 0 or μm = 1, 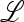 ex is a diagonal matrix, and its eigenvalues are
ex is a diagonal matrix, and its eigenvalues are
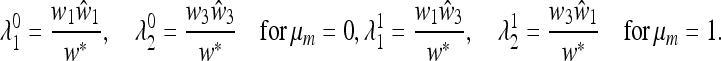 |
(31) |
The magnitude of the leading eigenvalue depends on the asymmetry of the parameters 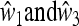 , with respect to w1 and w3. This asymmetry may be represented by the parameter
, with respect to w1 and w3. This asymmetry may be represented by the parameter
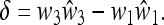 |
(32) |
As the fitnesses are relative, and hence determined up to a multiplicative positive constant in each generation, δ = 0 implies that 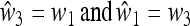 , namely complete symmetry between the two alleles A and a, whereas δ ≠ 0 implies asymmetry.
, namely complete symmetry between the two alleles A and a, whereas δ ≠ 0 implies asymmetry.
When δ = 0 (i.e., the symmetric case 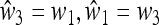 discussed in the next section), it turns out that
discussed in the next section), it turns out that 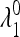 and
and 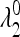 are <1, whereas at least one of the eigenvalues
are <1, whereas at least one of the eigenvalues 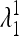 and
and 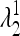 is >1. Therefore when μm = 0,
is >1. Therefore when μm = 0, 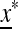 is externally stable for all 0 < μM ≤ 1, and when μm = 1,
is externally stable for all 0 < μM ≤ 1, and when μm = 1, 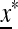 is never stable when 0 ≤ μM < 1. Thus when r = 0, a mutation-reducing allele cannot invade. The results are sensitive to the asymmetry measure δ, as is discussed in greater detail later.
is never stable when 0 ≤ μM < 1. Thus when r = 0, a mutation-reducing allele cannot invade. The results are sensitive to the asymmetry measure δ, as is discussed in greater detail later.
THE SYMMETRIC CASE WITH PERIOD 2
In the symmetric case where the selection regime alternates each generation, we have 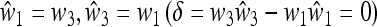 , and we can present a complete external stability analysis of the equilibrium
, and we can present a complete external stability analysis of the equilibrium 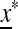 . In the symmetric case, the fitness parameters fluctuate between generations such that
. In the symmetric case, the fitness parameters fluctuate between generations such that
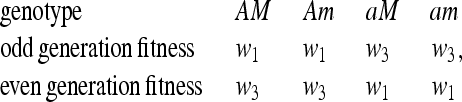 |
(33) |
where, of course, we assume w1 ≠ w3. The unique internally stable equilibrium 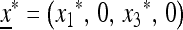 on the boundary where only allele M is present can be represented as
on the boundary where only allele M is present can be represented as 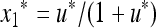 ,
, 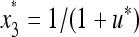 , where u* is the unique positive root of the quadratic Equation 15, which becomes
, where u* is the unique positive root of the quadratic Equation 15, which becomes
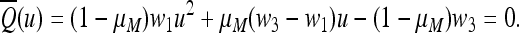 |
(34) |
In File S3 we use (34) to analyze the factors in (29). From (29), M(1) = 0 when r = 1, in which case the larger eigenvalue is 1. When 0 ≤ r < 1, the sign of M (1) depends on the signs of (μm − μM) and the sign of Δ = Δ (r, μm). In fact we show in File S3 that the bilinear function Δ(r, μm) is always negative in the symmetric case. As an immediate consequence of this, together with the representation of M(1) given in (29), we secure the following result.
Result 2. The internally stable equilibrium is externally stable when μM > μm and is unstable when μM < μm for all 0 ≤ r < 1. Thus with symmetric changing environments, higher mutation rates are favored.
As in the case of constant environments, Result 2 accords with the behavior of the mean fitness at equilibrium as a function of the resident mutation rate. With symmetric changing environments we can show the following.
Result 3. The mean fitness w* = w*(μM) at equilibrium is a monotone increasing function of the mutation rate μM.
The proof of this result can be found in File S4.
THE SYMMETRIC CASE WITH PERIOD 4
The fitnesses of the genotypes can be represented as follows:
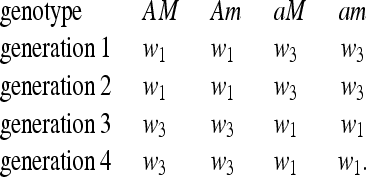 |
(35) |
In this case, on the boundary where M is fixed, the four-generation recursion is
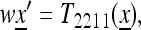 |
(36) |
where, as in (21) and (22),
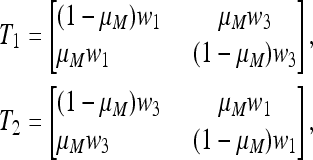 |
(37) |
and the four-generation normalization factor is then
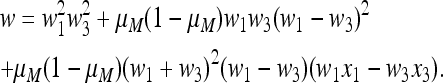 |
(38) |
Our argument proceeds in much the same way as for the symmetric model with period 2. First we show in File S5, Part I that when r = 0,
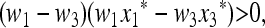 |
(39) |
where 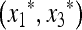 denotes the equilibrium of the four-generation transformation. Hence, from (38)
denotes the equilibrium of the four-generation transformation. Hence, from (38)
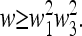 |
(40) |
We now turn to the external stability of 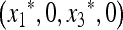 , which is determined by the eigenvalues of
, which is determined by the eigenvalues of 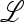 ex. When μM = 0, we can write
ex. When μM = 0, we can write
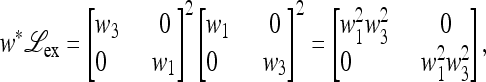 |
(41) |
and when μM = 1,
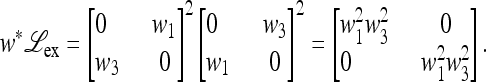 |
(42) |
But by (40), 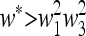 , and therefore
, and therefore 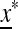 is externally stable when either μM = 0 or μM = 1. In other words, if 0 < μM < 1, then a new mutation-modifying allele cannot invade
is externally stable when either μM = 0 or μM = 1. In other words, if 0 < μM < 1, then a new mutation-modifying allele cannot invade 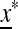 if it produces mutation rates that are too high or too low. This leaves open the possibility that there might be an intermediate value of the mutation rate μM that gives rise to an equilibrium
if it produces mutation rates that are too high or too low. This leaves open the possibility that there might be an intermediate value of the mutation rate μM that gives rise to an equilibrium 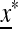 on the M-fixed boundary that cannot be invaded. The first indication that
on the M-fixed boundary that cannot be invaded. The first indication that 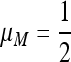 is this value is suggested by the following.
is this value is suggested by the following.
Result 4. In the period 4 symmetric case, if r = 0, the mean fitness w* = w*(μM) achieves a maximum at 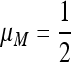 .
.
The proof is in File S5, Part II.
If 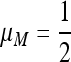 , then the single-generation transformations are very symmetric, and the equilibrium on the M-fixed boundary has
, then the single-generation transformations are very symmetric, and the equilibrium on the M-fixed boundary has 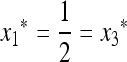 . To examine the invasion by
. To examine the invasion by 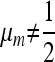 we evaluate the external stability of
we evaluate the external stability of 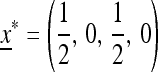 . The eigenvalues of the linear approximation
. The eigenvalues of the linear approximation 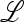 ex near
ex near 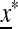 are the roots of M (z) = 0, where M (z) can be expressed as
are the roots of M (z) = 0, where M (z) can be expressed as
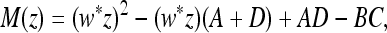 |
(43) |
where the entries of 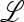 ex, A, B, C, D are all positive. As before, one eigenvalue is positive, and it can be shown that
ex, A, B, C, D are all positive. As before, one eigenvalue is positive, and it can be shown that 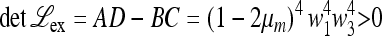 . Hence both eigenvalues are positive. The larger one is <1 if M(1) > 0, which is shown to be the case in File S5, Parts III and IV. This allows us to claim the following.
. Hence both eigenvalues are positive. The larger one is <1 if M(1) > 0, which is shown to be the case in File S5, Parts III and IV. This allows us to claim the following.
Result 5. In the period 4 symmetric case with 0 ≤ r ≤ 1, the mutation rate 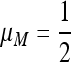 cannot be invaded by mutation rates either > or <
cannot be invaded by mutation rates either > or <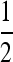 .
.
For cycles of period m = 2n, when r = 0, we can show that if 0 < μM ≤ 1, a modifier allele m that produces μm = 0 cannot invade. If n is even we are also able to prove that allele m with μm = 1 cannot invade. It remains to prove the latter result for odd values of n, and we see in numerical analysis that this could be difficult.
THE MEAN FITNESS AND EIGENVALUE CRITERIA
The analyses by Ishii et al. (1989) and Gaál et al. (2010) formulated the dynamics of M and m, the mutation-controlling alleles, in terms of the numbers of each genotype. In particular, when r = 0, Ishii et al. concluded that the criterion for invasion and long-term increase of m in a population monomorphic for M could be obtained from the limiting geometric mean of the mean fitnesses at each generation. This allowed them to state that when r = 0, the uninvadable mutation rate “maximizes the long-term geometric average of population fitness” (Ishii et al. 1989, p. 165). This harks back to early analyses on neutral modifiers of recombination and the mean fitness principle of Karlin and McGregor (1972, 1974), who hypothesized that uninvadable values of neutral modifiers of recombination, mutation, and migration also maximized the mean fitness.
Our criterion for initial increase is in terms of the local stability of the equilibrium 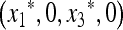 , where
, where 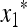 and
and 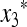 are functions of μM, to invasion by m. This invasion is determined by the leading eigenvalue λ of the local stability matrix that gives the frequencies of Am and am near
are functions of μM, to invasion by m. This invasion is determined by the leading eigenvalue λ of the local stability matrix that gives the frequencies of Am and am near 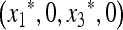 , and λ must be a function of both μm and μM. As pointed out above, the relevant properties of λ are expressible in terms of properties of M(λ), the characteristic polynomial of the local stability matrix
, and λ must be a function of both μm and μM. As pointed out above, the relevant properties of λ are expressible in terms of properties of M(λ), the characteristic polynomial of the local stability matrix 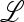 ex. The value of μm that makes
ex. The value of μm that makes 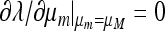 will be the stable value of the mutation rate.
will be the stable value of the mutation rate.
We write the local stability matrix for local dynamics of Am, am near 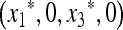 as
as
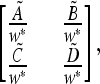 |
(44) |
where the entries 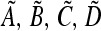 are defined by the continued operation of the linear fractional transformation corresponding to
are defined by the continued operation of the linear fractional transformation corresponding to 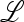 ex, w* is a function of μM, and the tilde indicates that the numerators of all entries are functions of μm. We can write the characteristic polynomial M(λ) of matrix (44) as
ex, w* is a function of μM, and the tilde indicates that the numerators of all entries are functions of μm. We can write the characteristic polynomial M(λ) of matrix (44) as
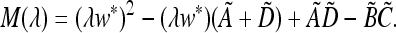 |
(45) |
From (45) we can calculate ∂λ/∂μm.
For the initial equilibrium 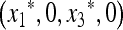 , where allele m is absent, we have
, where allele m is absent, we have
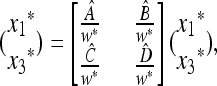 |
(46) |
where the circumflex indicates that the numerators are functions of μM. From (46), the matrix has an eigenvalue of 1, and we have
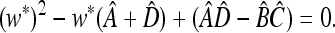 |
(47) |
The stationary values of w* with respect to μM can be obtained by differentiating Equation 47 with respect to μM and finding values of μM such that ∂w*/∂μM = 0. The relationship between mutation values, which maximize the mean fitness, and the uninvadable value of μ that sets 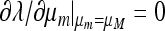 is given by the following.
is given by the following.
Result 6. Mutation rates μM that are critical points of the mean fitness w* in the case r = 0 also entail that at μm = μM, ∂λ/∂μm = 0.
The proof is in the appendix. Figure 1 plots ∂w*/∂μM and ∂λ/∂μm for n = 3, and the two derivatives are seen to vanish at the same value of the mutation rate. The proof of Result 6 actually makes no use of the symmetry of the fitnesses, so Result 6 is also true for asymmetric fitnesses.
NUMERICAL ANALYSIS
Result 2 entails that in the symmetric case 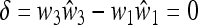 , for period 2 (n = 1) for all values of the recombination rate, nonzero values of the resident mutation rate μM will be invaded by allele m that gives rise to a higher mutation rate; i.e., μm > μM. The mutation rate will therefore go on increasing to its maximum of 1. A similar result for period 4 (n = 2), namely Result 5, tells us that for all r, values of
, for period 2 (n = 1) for all values of the recombination rate, nonzero values of the resident mutation rate μM will be invaded by allele m that gives rise to a higher mutation rate; i.e., μm > μM. The mutation rate will therefore go on increasing to its maximum of 1. A similar result for period 4 (n = 2), namely Result 5, tells us that for all r, values of 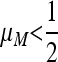 will be invaded by higher values μm > μM, but values of
will be invaded by higher values μm > μM, but values of 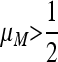 will be invaded by lower values of μm. There are no constraints on the strength of the symmetric selection as were required in the analysis by Ishii et al. (1989). There have been suggestions in the literature that the stability of the mutation rate μ = 1/n for the symmetric model cycles with period 2n holds for larger values of n (Ishii et al. 1989; Lachmann and Jablonka 1996). For the asymmetric model, with δ ≠ 0, however, Salathé et al. (2009) showed that the situation was much more complicated and that if the asymmetry was large enough, zero was the stable mutation rate; mutation provided no advantage with fixed or random period cycles. Further, with stochastic environments and a rate of environmental change of 1/n every generation, the stable switching rate can be up to two orders of magnitude lower than 1/n (Salathé et al. 2009, Figure 1). In addition, we noted after Result 5 above that even in the case r = 0, if n is odd there may be some anomalies.
will be invaded by lower values of μm. There are no constraints on the strength of the symmetric selection as were required in the analysis by Ishii et al. (1989). There have been suggestions in the literature that the stability of the mutation rate μ = 1/n for the symmetric model cycles with period 2n holds for larger values of n (Ishii et al. 1989; Lachmann and Jablonka 1996). For the asymmetric model, with δ ≠ 0, however, Salathé et al. (2009) showed that the situation was much more complicated and that if the asymmetry was large enough, zero was the stable mutation rate; mutation provided no advantage with fixed or random period cycles. Further, with stochastic environments and a rate of environmental change of 1/n every generation, the stable switching rate can be up to two orders of magnitude lower than 1/n (Salathé et al. 2009, Figure 1). In addition, we noted after Result 5 above that even in the case r = 0, if n is odd there may be some anomalies.
We have therefore explored three effects numerically: the role of n and in particular whether it is odd or even, the role of asymmetry in the cycling fitness values (extending Salathé et al. 2009), and the effect of recombination between the major locus and the mutation modifier. The equilibrium 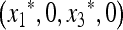 is found numerically and for given parameter values, the matrix
is found numerically and for given parameter values, the matrix  ex is determined numerically together with its eigenvalues. The leading eigenvalue λ1 is numerically differentiated with respect to μm and the derivative evaluated at μM = μm, where as we showed in (29) the eigenvalue is 1. The sign of
ex is determined numerically together with its eigenvalues. The leading eigenvalue λ1 is numerically differentiated with respect to μm and the derivative evaluated at μM = μm, where as we showed in (29) the eigenvalue is 1. The sign of 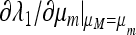 then tells us whether a larger or smaller value of μm will produce λ1 > 1 and hence cause m to invade. Thus, points where ∂λ1/∂μ crosses zero with a negative slope indicate values of μM that cannot be invaded by any nearby value of μm. These values of μM can be regarded as evolutionarily stable. These results are summarized in Figure 2.
then tells us whether a larger or smaller value of μm will produce λ1 > 1 and hence cause m to invade. Thus, points where ∂λ1/∂μ crosses zero with a negative slope indicate values of μM that cannot be invaded by any nearby value of μm. These values of μM can be regarded as evolutionarily stable. These results are summarized in Figure 2.
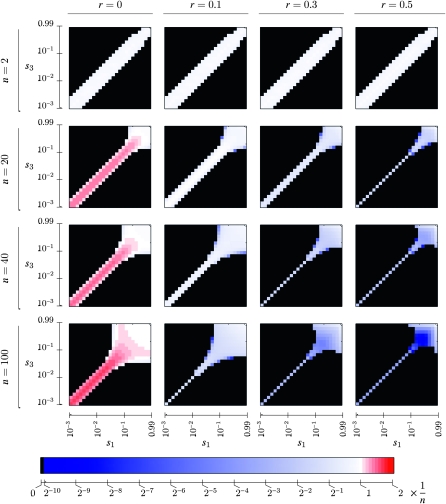
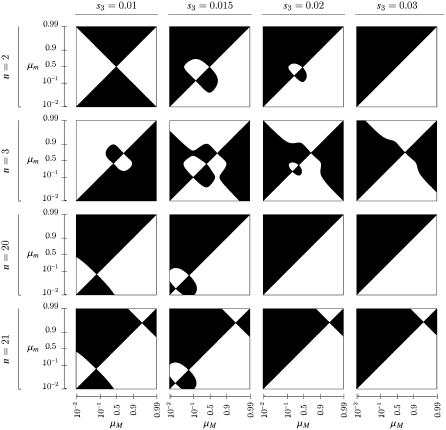
Figure 2.—
Evolutionarily stable mutation rates as a function of the selection coefficients for the disfavored allele in each environment, s1 = 1 − w1 and s3 = 1 − w3, which are plotted logarithmically. The number of generations in each environment, n, is given at the left. The recombination rate between the major locus under selection and the modifier locus is given at the top. The colors denote the stable mutation rate and are given in units of the canonical predicted rate of 1/n in the colorbar at the bottom; white is a rate of 1/n, shades of red are rates >1/n, and blues are rates <1/n. Black denotes evolution to zero mutation rate.
Figure 2 extends Figure 2 of Salathé et al. (2009) by exhibiting the roles of recombination and asymmetry on the stable mutation rate. We point out first that the first row of graphs in Figure 2 represents results for n = 2 (i.e., period 4 case) and we see that with perfect symmetry in the cycling fitnesses, the mutation rate 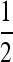 is stable for all recombination rates, verifying Result 5 for all r. In what follows, we use the notation
is stable for all recombination rates, verifying Result 5 for all r. In what follows, we use the notation 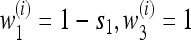 for n generations with
for n generations with 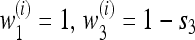 for the next n generations. Thus, s1 = s3 corresponds to perfect symmetry in selection regimes (Salathé et al. 2009). Note that in all graphs with r = 0 and in many of the others, there is an interval of asymmetry in the selection coefficients s1 = 1 − w1 and s3 = 1 − w3 in which there is a stable nonzero mutation rate. As n and r increase, the stable nonzero mutation rate with s1 = s3 decreases. For r = 0 and small s1 and s3, the extent of asymmetry that allows a stable nonzero mutation rate does not change much with n, whereas for the r > 0 values shown in Figure 2 it is very sensitive to n.
for the next n generations. Thus, s1 = s3 corresponds to perfect symmetry in selection regimes (Salathé et al. 2009). Note that in all graphs with r = 0 and in many of the others, there is an interval of asymmetry in the selection coefficients s1 = 1 − w1 and s3 = 1 − w3 in which there is a stable nonzero mutation rate. As n and r increase, the stable nonzero mutation rate with s1 = s3 decreases. For r = 0 and small s1 and s3, the extent of asymmetry that allows a stable nonzero mutation rate does not change much with n, whereas for the r > 0 values shown in Figure 2 it is very sensitive to n.
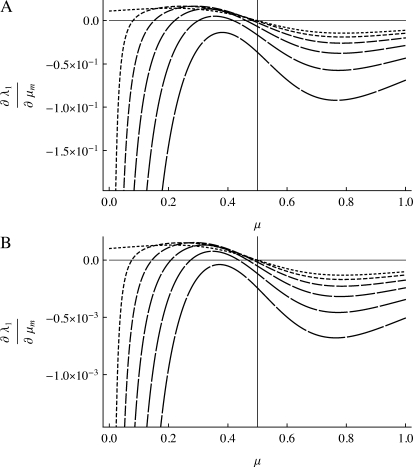
To ascertain that the existence of a range of asymmetry permitting a stable nonzero mutation rate was not an artifact of the numerical procedure, for r = 0 the n = 2 and n = 3 cases were repeated using a very accurate numerical derivative [complex step derivative (Squire and Trapp 1998)] and a finer array of s3 values for s1 = 0.01 and s1 = 0.1. The n = 2 results are in Figure 3 where there can be no doubt that this band of asymmetric fitnesses allowing a stable mutation rate at or very close to 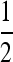 really exists. We see that in Figure 3, A and B, ∂λ1/∂μ is positive for
really exists. We see that in Figure 3, A and B, ∂λ1/∂μ is positive for 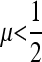 and negative for
and negative for 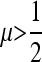 when s1 = s3 and that μ-values near
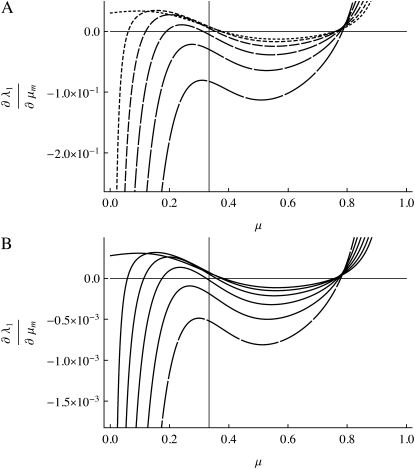
when s1 = s3 and that μ-values near 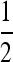 are stable for a range of μ-values that decreases as the asymmetry increases, with μ = 0 becoming stable when the asymmetry is too great. The width of the band of stability is a little <1 log10 fitness unit in both Figures 3 and 4. Figure 4 shows the same effect for n = 3. In Figure 4, A and B, ∂λ1/∂μm = 0 occurs close to
are stable for a range of μ-values that decreases as the asymmetry increases, with μ = 0 becoming stable when the asymmetry is too great. The width of the band of stability is a little <1 log10 fitness unit in both Figures 3 and 4. Figure 4 shows the same effect for n = 3. In Figure 4, A and B, ∂λ1/∂μm = 0 occurs close to 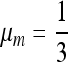 , but not exactly at
, but not exactly at 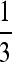 , which would be the case if 1/n were the evolutionarily stable mutation rate.
, which would be the case if 1/n were the evolutionarily stable mutation rate.
Figure 3.—
Plot of the derivative of the leading eigenvalue, λ1, with respect to μm evaluated at μM = μm = μ when n = 2 and r = 0. (A) s1 = 1 − w1 = 10−1 and the lines from shortest dashes to longest dashes are s3 = 1 − w3 = 10−1, 10−0.9, 10−0.8, 10−0.7, 10−0.6, and 10−0.5. (B) s1 = 0.01 and the lines from shortest dashes to longest dashes are s3 = 10−2, 10−1.9, 10−1.8, 10−1.7, 10−1.6, and 10−1.5. The vertical line is located at μ = 1/n = 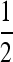 .
.
Figure 4.—
(A and B) The same as Figure 3, except n = 3. The vertical line is located at μ = 1/n = 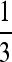 .
.
Figures 5 and 6 offer another view of the stability/invasibility parameter ranges. Figure 5 sets r = 0 while r = 0.3 in Figure 6. In these figures, the solid areas have the leading eigenvalue <1, while it is >1 in the open regions. In the first row of both figures we see first the stability of 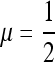 , and as s3 increases the black area under the separating diagonal reflects the range of stable mutation rates near
, and as s3 increases the black area under the separating diagonal reflects the range of stable mutation rates near 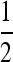 , which disappears when s1 = 0.01 and s1 = 0.03, leaving zero as the stable mutation rate. In general, we see that when s3 is different from s1, an unstable equilibrium in μ appears below the stable equilibrium. If the mutation changes in sufficiently small steps, then μ can be trapped below the unstable equilibrium and evolve to 0. As long as the rate may take larger steps, and if the rate remains below
, which disappears when s1 = 0.01 and s1 = 0.03, leaving zero as the stable mutation rate. In general, we see that when s3 is different from s1, an unstable equilibrium in μ appears below the stable equilibrium. If the mutation changes in sufficiently small steps, then μ can be trapped below the unstable equilibrium and evolve to 0. As long as the rate may take larger steps, and if the rate remains below 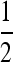 , it will eventually converge to the stable equilibrium. As s3 increases further, the stable and unstable equilibria annihilate one another and μ goes to 0, if it remains below
, it will eventually converge to the stable equilibrium. As s3 increases further, the stable and unstable equilibria annihilate one another and μ goes to 0, if it remains below 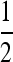 .
.
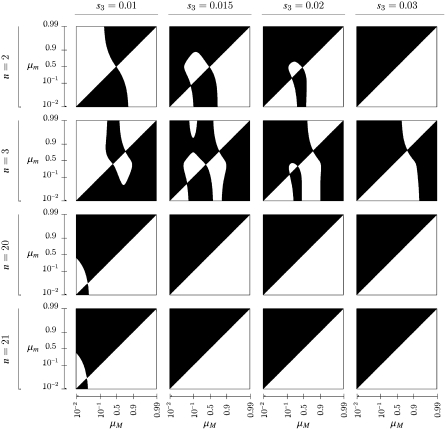
Figure 5.—
Pairwise invasibility plots for mutation rate modifier. Combinations of resident mutation rate μM and invading mutation rate μm in the open regions yield a leading eigenvalue >1 in absolute magnitude; the leading eigenvalue is <1 in the solid regions. We assume that w1 = 1 − s1 and w3 = 1 − s3 and set s1 = 0.01. Each row denotes a different value for n, the number of generations in each environment. The recombination rate is set to r = 0. Note that the x- and y-axes are logarithmically scaled from 10−2 to 0.5 and from 1 − 10−2 to 1 − 0.5.
Figure 6.—
The same as Figure 5 except the recombination rate is set to r = 0.3.
With n odd, the complication alluded to after Result 5 becomes visible: the first columns of Figures 5 and 6 show the symmetric model, and the second and fourth rows are examples with n odd. With n = 3 we see a stable value of the mutation rate <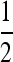 and near
and near 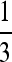 . In general, when n is odd, and r = 0, an unstable equilibrium value of μ appears with
. In general, when n is odd, and r = 0, an unstable equilibrium value of μ appears with 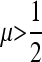 . Thus, larger values of μ may also be invadable by even larger values, depending on the difference between the resident μM and the invading μm. The domains of attraction in the asymmetric cases can become very complicated; compare s3 = 0.015, n = 3, and r = 0 with r = 0.3. Note that with n = 20 and 21, r = 0.3 forces stability of μ = 0 for most cases, while with r = 0, s3 = 0.015, and n = 21, there is a range of high mutation rates that will be invaded by even higher (although unrealistic) mutation rates. Examination of Figures 5 and 6 suggests that if both mutation rates μM and μm are very small, odd and even values of n give qualitatively similar results. For values of μM and μm <
. Thus, larger values of μ may also be invadable by even larger values, depending on the difference between the resident μM and the invading μm. The domains of attraction in the asymmetric cases can become very complicated; compare s3 = 0.015, n = 3, and r = 0 with r = 0.3. Note that with n = 20 and 21, r = 0.3 forces stability of μ = 0 for most cases, while with r = 0, s3 = 0.015, and n = 21, there is a range of high mutation rates that will be invaded by even higher (although unrealistic) mutation rates. Examination of Figures 5 and 6 suggests that if both mutation rates μM and μm are very small, odd and even values of n give qualitatively similar results. For values of μM and μm < 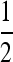 , the patterns for n = 2 and n = 3 are similar for both r = 0 and r = 0.3. In the unrealistic range of mutation rates >0.5, n = 2 and n = 3 show different patterns of invasibility with μ = 1 stable for n = 3 but not for n = 2.
, the patterns for n = 2 and n = 3 are similar for both r = 0 and r = 0.3. In the unrealistic range of mutation rates >0.5, n = 2 and n = 3 show different patterns of invasibility with μ = 1 stable for n = 3 but not for n = 2.
For large values of s1 and s3 the range of asymmetry that allows a stable nonzero mutation rate increases as n increases, but for given n, it decreases as r increases. Note that for larger recombination rates and larger n, the stable mutation rates, if different from 0, are ≪1/n. Comparing the effect of recombination when n is small with that when n is large, we have an indication from Figure 5 (r = 0) and Figure 6 (r = 0.3) that increasing recombination makes it more likely that the mutation rate will decrease.
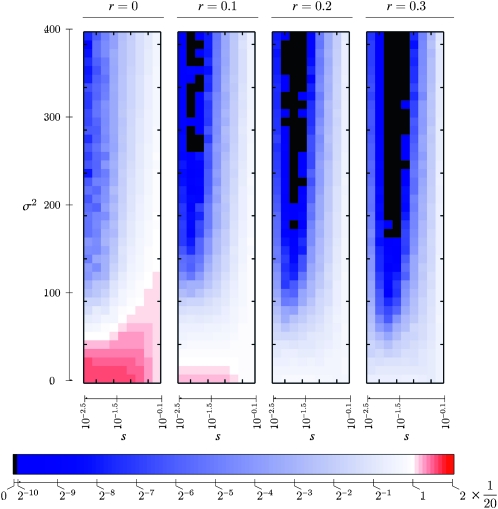
Salathé et al. (2009) showed for the symmetric selection case that if the period of the cycle was a gamma-distributed random variable, the stable switching rate dropped precipitously as the variance in the cycle period increased, compared to the case where the period was fixed at the mean of the gamma distribution. Figure 7 illustrates the interaction between recombination and variance in the cycle period. The effect of recombination on the evolutionarily stable mutation rate is striking: with r = 0.2, for example (second panel from the right in Figure 7), none of the mutation rates near 1/n that are stable when the period is fixed at 2n (red–pink in the leftmost panel) are stable. Moving from left to right in Figure 7, recombination is seen to strengthen the effect of randomness in the period seen by Salathé et al. (2009, Figure 1) in shifting the stable mutation rate to substantially <1/n. It is reasonable to expect that with asymmetries in fitness, recombination would also enhance the tendency of variance in the cycle period to decrease the evolutionarily stable rate of phenotypic switching.
Figure 7.—
Simulated evolutionarily stable mutation rates with w1 = w3 = 1 − s. The numbers of generations in each environment are drawn from a gamma distribution with mean 20 and variance σ2. Each panel denotes a different value of the recombination rate given in the first row. Colors denote the stable mutation rate, as in Figure 2. Each stable rate is the average of 100 simulations; each simulation consists of iterating the recursion system (8) to determine whether a new rate μm can invade an old rate μM. One thousand successive new rates are sequentially assayed for whether they invade; the new rate invades and replaces the old rate if it increases by 1.001 over its initial frequency after 200,000 generations. More on the simulation methods can be found in Salathé et al. (2009).
DISCUSSION
The contribution of epigenetic phenomena to both statistical and dynamic properties of phenotypic variation has received much recent attention. In general, the two aspects, statistical and dynamic, have been well separated. The statistical treatment by Slatkin (2009) quantifies the contribution of epigenetic change to disease risk, where the epigenetic effect could be mutational and act on gene expression. Another recent analysis by Danchin and Wagner (2010) introduces transmitted epigenetic variance as well as “culture” through transmitted social variance into calculations of heritability; addition of these effect leads them to “inclusive heritability,” which they define as all inherited components of phenotypic variance. Their framework is a special case of early work on the contributions of cultural inheritance to estimated heritability by Cavalli-Sforza and Feldman (1978) and Feldman and Cavalli-Sforza (1979) that predates the current interest in cultural inheritance as a possible cause of transgenerational epigenetics.
Such statistical treatments usually do not include recombination as a force that affects the genetic or epigenetic contributions to phenotypic variance. For example, multilocus evolutionary simulation studies by Feinberg and Irizarry (2010) of epigenetic variation were stimulated by findings of dietary modification of DNA methylation of the Agouti gene in mice and methylation of the Axin-fused allele in kinked-tail mice (Cooney et al. 2002; Rakyan et al. 2003; Waterland and Jirtle 2003). Recombination was not included in the simulations by Feinberg and Irizarry.
Studies motivated by the adjustment by microorganisms of their gene regulation to environmental change, such as those mentioned in the Introduction, ignore recombination because it is assumed to be rare in the bacteria, yeast, or cellular populations on which the models are based. As a general rule, these models take populations of cells that do not compete and compare their exponential growth dynamics under different conditions. Thus, the recent study by Leibler and Kussell (2010) focused on the historical phenotypic states of a population of nonrecombining cells, and fitness in different populations was measured as a property of these histories. Recombination would be difficult to include in such a treatment for the same reason that coalescent analysis in genetics is complicated by recombination and selection. Similarly Gerland and Hwa (2009) measure the success in populations of cells subject to mutation by the genetic load, namely the reduction below the maximum fitness possible caused by deleterious mutations in a single population. Here again the analysis is in terms of a single gene, and recombination does not enter into the dynamics of the different mutational states. However, Lynch (2007, p. 89) points out that “recombination at the nucleotide level does not appear to be exceptionally low in prokaryotes when compared to that in multicellular species.”
Inclusion of recombination between the locus under selection and the locus that controls the mutation rate is important but greatly increases the difficulty of exact analysis when the selection fluctuates over time. Formally the reason for this is that at r = 0 the recursion system is linear fractional, and iterates of linear fractional transformations remain linear fractional, even if the parameters are not constant over time. In particular, this guarantees that we can model the starting point of the modification process as the unique stable trajectory of the compound linear fractional system. The initial dynamics of the rare mutation-modifying allele can then be analyzed as in Liberman and Feldman (1986). The argument makes use of the positivity of the initial increase matrix specified by 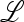 ex (Equations 17–19). When r > 0, the linear fractional structure is replaced by a much more complicated dynamical iteration whose properties are difficult to discern in general. With sufficiently weak selection and small values of
ex (Equations 17–19). When r > 0, the linear fractional structure is replaced by a much more complicated dynamical iteration whose properties are difficult to discern in general. With sufficiently weak selection and small values of 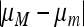 , however, M'Gonigle et al. (2009) made considerable progress in the case where cycling was due to host–parasite interactions.
, however, M'Gonigle et al. (2009) made considerable progress in the case where cycling was due to host–parasite interactions.
Our analysis of the symmetric cases with n = 1 and 2, corresponding to periods 2 and 4, respectively, for the fitness cycle, confirms results found numerically by Ishii et al. (1989). They were able to study the system analytically in these cases under one of two restrictive conditions: the weak selection limit 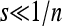 (where, in our terminology, s1 = 1 − w1, for example, as in Figures 1 through 7) or s1 close to 1 (which entails in our terminology that 1 − w1 ≈ 0). Our Results 3 and 5 and the proofs in File S1, File S2, File S3, File S4, and File S5 show analytically that μ = 1 and
(where, in our terminology, s1 = 1 − w1, for example, as in Figures 1 through 7) or s1 close to 1 (which entails in our terminology that 1 − w1 ≈ 0). Our Results 3 and 5 and the proofs in File S1, File S2, File S3, File S4, and File S5 show analytically that μ = 1 and 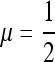 are the uninvadable mutation rates for n = 1 and n = 2, respectively, with no restrictions on the recombination rate or fitnesses w1 and w3.
are the uninvadable mutation rates for n = 1 and n = 2, respectively, with no restrictions on the recombination rate or fitnesses w1 and w3.
As pointed out by Ishii et al. (1989), odd values of n produce two equilibria for the mutation rate, a stable value <0.5 and an unstable one >0.5. As n becomes larger, however, our Figures 5 and 6, with n = 21 and symmetric fitnesses show that increasing r can eliminate the large unstable value of μ; with r = 0.3, in the first column of Figure 6, the value of 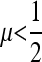 is stable from above μ, while in Figure 5, where r = 0, μ = 1 has a domain of stability above the unstable equilibrium.
is stable from above μ, while in Figure 5, where r = 0, μ = 1 has a domain of stability above the unstable equilibrium.
In both symmetric cases n = 1 and n = 2, we proved that the mutation rates that cannot be invaded by any other (μ = 1 for n = 1 and 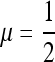 for n = 2) are also the mutation rates that maximize the mean fitness w*. Those studies (e.g., Ishii et al. 1989; Gaál et al. 2010) that argue in terms of long-term growth rates in exponentially growing populations find that this is true for any n. Our Result 5 provides an elegant resolution of the difference, with symmetric or asymmetric fitnesses and r = 0: the value of μ where ∂λ/∂μ vanishes, namely the value that gives “eigenvalue stability,” is the same value that maximizes the logarithm of the long-term mean fitness, namely
for n = 2) are also the mutation rates that maximize the mean fitness w*. Those studies (e.g., Ishii et al. 1989; Gaál et al. 2010) that argue in terms of long-term growth rates in exponentially growing populations find that this is true for any n. Our Result 5 provides an elegant resolution of the difference, with symmetric or asymmetric fitnesses and r = 0: the value of μ where ∂λ/∂μ vanishes, namely the value that gives “eigenvalue stability,” is the same value that maximizes the logarithm of the long-term mean fitness, namely 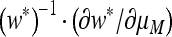 . Numerical analysis confirms that
. Numerical analysis confirms that 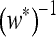 is indeed the factor that relates these derivatives, and the proof is given in the appendix.
is indeed the factor that relates these derivatives, and the proof is given in the appendix.
Our analysis of symmetric selection shows some degree of robustness. From Figure 2 (and also Figures 3 and 4), there is a band of mildly asymmetric fitnesses surrounding the symmetric selection values s1 = 1 − w1 = s3 = 1 − w3 in which the stable mutation rate is very close indeed to that obtained under symmetric fitnesses. For large values of both s1 and s3 this band becomes wider as n increases for all recombination values (top right of each panel in Figure 2). For weaker asymmetric selection this band narrows substantially, and the stable mutation rate becomes smaller as the recombination rate increases. Thus, as r increases, a greater range of fitness asymmetries leads to a stable mutation rate of zero, except for n = 2. Even for n = 3 in the symmetric case, however, Figure 4 shows clear distance between the stable values of μ and the value 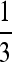 predicted, e.g., by Ishii et al. (1989) and Lachmann and Jablonka (1996). At n = 100, in Figure 2, the stable μ in the symmetric case is much closer to
predicted, e.g., by Ishii et al. (1989) and Lachmann and Jablonka (1996). At n = 100, in Figure 2, the stable μ in the symmetric case is much closer to 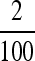 than to
than to 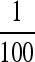 . As r increases, the stable value of μ in the symmetric case rapidly decreases below 1/n, and the band of stable nonzero mutation rates disappears. These results suggest that the optimal rate of phenotypic switching depends as much on the strength of selection in each environment as it does on the frequency of environmental changes, represented here by n; Kussell et al. (2005) claimed that n was much more important than the magnitudes of s1 and s3 in determining the uninvadable μ. At least for the values of n we tried, Figures 2, 5, and 6 show that s1 and s3 are indeed important.
. As r increases, the stable value of μ in the symmetric case rapidly decreases below 1/n, and the band of stable nonzero mutation rates disappears. These results suggest that the optimal rate of phenotypic switching depends as much on the strength of selection in each environment as it does on the frequency of environmental changes, represented here by n; Kussell et al. (2005) claimed that n was much more important than the magnitudes of s1 and s3 in determining the uninvadable μ. At least for the values of n we tried, Figures 2, 5, and 6 show that s1 and s3 are indeed important.
Our earlier numerical study (Salathé et al. 2009) showed that in asymmetric fitness landscapes (w1 ≠ w3), stable nonzero mutation rates required very strong selection in both environments (i.e., large s1 and s3). Our Figures 5 and 6 show an interesting feature of the dynamical system that distinguishes odd and even values of n. The numerical analysis illustrates that there may be more than one solution to 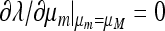 and that the direction of selection on μm might depend delicately on the sign and magnitude of (μM − μm). Why this is more likely to happen when n is odd than when n is even remains an interesting mathematical question. In both Figures 5 and 6, i.e., recombination r = 0.0 and 0.3, for n = 3, there are three critical values of μ when s1 = 0.01 and s3 = 0.015 and 0.02; one value is >0.5 and unstable, and two values are <0.5, with the greater of these locally stable, while the smaller one is locally unstable. The same pattern is exhibited for n = 21 with r = 0 (Figure 5), but not for r = 0.3 (Figure 6). Figures 2 and 7 suggest that increasing recombination makes it less likely that a stable nonzero mutation rate will exist with either a fixed or a random switching period (see also M'Gonigle et al. 2009). This suggests that switching in sexual organisms is less likely than in asexuals, and that if it occurs by genetic regulation of gene expression, the regulators should be linked to the genes they regulate. The role of recombination becomes more important as the period becomes longer, especially in symmetric or weakly asymmetric selection regimes.
and that the direction of selection on μm might depend delicately on the sign and magnitude of (μM − μm). Why this is more likely to happen when n is odd than when n is even remains an interesting mathematical question. In both Figures 5 and 6, i.e., recombination r = 0.0 and 0.3, for n = 3, there are three critical values of μ when s1 = 0.01 and s3 = 0.015 and 0.02; one value is >0.5 and unstable, and two values are <0.5, with the greater of these locally stable, while the smaller one is locally unstable. The same pattern is exhibited for n = 21 with r = 0 (Figure 5), but not for r = 0.3 (Figure 6). Figures 2 and 7 suggest that increasing recombination makes it less likely that a stable nonzero mutation rate will exist with either a fixed or a random switching period (see also M'Gonigle et al. 2009). This suggests that switching in sexual organisms is less likely than in asexuals, and that if it occurs by genetic regulation of gene expression, the regulators should be linked to the genes they regulate. The role of recombination becomes more important as the period becomes longer, especially in symmetric or weakly asymmetric selection regimes.
Acknowledgments
The authors thank Freddy Christiansen for his careful reading of an earlier draft and two anonymous referees for incisive and useful comments. This research was supported in part by National Institutes of Health grant GM 28016.
APPENDIX
Proof that 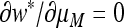 if and only if
if and only if 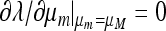 .
.
From (45) we have at λ = 1,
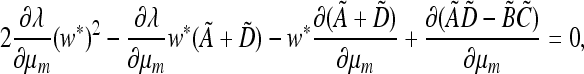 |
(A1) |
which we can reorganize at μm = μM as
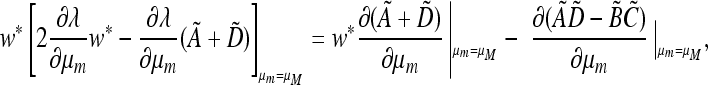 |
(A2) |
or
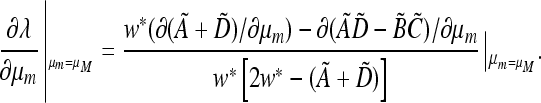 |
(A3) |
But, from Equation 47,
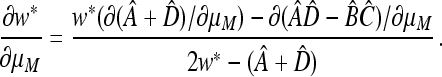 |
(A4) |
Since 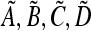 are exactly the same as
are exactly the same as 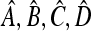 with μm replacing μM, we conclude that
with μm replacing μM, we conclude that
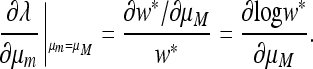 |
(A5) |
That is, 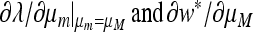 vanish together.
vanish together.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123620/DC1.
References
- Acar, M., J. T. Mettetal and A. van Oudenaarden, 2008. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 40 471–475. [DOI] [PubMed] [Google Scholar]
- Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik and S. Leibler, 2004. Bacterial persistence as a phenotypic switch. Science 305 1622–1625. [DOI] [PubMed] [Google Scholar]
- Bergman, A., and M. W. Feldman, 1990. More on selection for and against recombination. Theor. Popul. Biol. 38 68–92. [DOI] [PubMed] [Google Scholar]
- Bonduriansky, R., and T. Day, 2009. Nongenetic inheritance and its evolutionary implications. Annu. Rev. Ecol. Syst. 40 103–125. [Google Scholar]
- Cavalli-Sforza, L. L., and M. W. Feldman, 1978. The evolution of continuous variation. III. Joint transmission of genotype, phenotype and environment. Genetics 90 391–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1976. Recombination modification in a fluctuating environment. Genetics 83 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney, C. A., A. A. Dave and G. L. Wolff, 2002. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 132(Suppl. 8): 23935–24005. [DOI] [PubMed] [Google Scholar]
- Danchin, E., and R. H. Wagner, 2010. Inclusive heritability: combining genetic and nongenetic information to study animal culture. Oikos 119 210–218. [Google Scholar]
- Feinberg, A. P., and R. A. Irizarry, 2010. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl. Acad. Sci. USA 107(Suppl. 1): 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M. W., 1972. Selection for linkage modification: I. Random mating populations. Theor. Popul. Biol. 3 324–346. [DOI] [PubMed] [Google Scholar]
- Feldman, M. W., and L. L. Cavalli-Sforza, 1979. Aspects of variance and covariance analysis with cultural inheritance. Theor. Popul. Biol. 15 276–307. [DOI] [PubMed] [Google Scholar]
- Feldman, M. W., and U. Liberman, 1986. An evolutionary reduction principle for genetic modifiers. Proc. Natl. Acad. Sci. USA 83 4824–4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M. W., F. B. Christiansen and L. D. Brooks, 1980. Evolution of recombination in a constant environment. Proc. Natl. Acad. Sci. USA 77 4838–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M. W., S. P. Otto and F. B. Christiansen, 1997. Population genetic perspectives on the evolution of recombination. Annu. Rev. Genet. 30 261–295. [DOI] [PubMed] [Google Scholar]
- Gaál, B., J. W. Pitchford and A. J. Wood, 2010. Exact results for the evolution of stochastic switching in variable asymmetric environments. Genetics 184 1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandon, S., and S. P. Otto, 2007. The evolution of sex and recombination in response to abiotic or coevolutionary fluctuations in epistasis. Genetics 175 1835–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerland, U., and T. Hwa, 2009. Evolutionary selection between alternative modes of gene regulation. Proc. Natl. Acad. Sci. USA 106 8841–8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, W. D., 1980. Sex versus non-sex versus parasite. Oikos 35 282–290. [Google Scholar]
- Haraguchi, Y., and A. Sasaki, 1996. Host-parasite arms race in mutation modifications: indefinite escalation despite a heavy load? J. Theor. Biol. 183 121–137. [DOI] [PubMed] [Google Scholar]
- Henderson, I. R., and S. E. Jacobsen, 2007. Epigenetic inheritance in plants. Nature 447 418–424. [DOI] [PubMed] [Google Scholar]
- Ishii, K., H. Matsuda, Y. Iwasa and A. Sasaki, 1989. Evolutionary stable mutation rate in a periodically changing environment. Genetics 121 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin, S., and J. L. McGregor, 1972. The evolutionary development of modifier genes. Proc. Natl. Acad. Sci. USA 69 3611–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin, S., and J. L. McGregor, 1974. Towards a theory of the evolution of modifier genes. Theor. Popul. Biol. 5 59–105. [DOI] [PubMed] [Google Scholar]
- Kussell, E., and S. Leibler, 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309 2075–2078. [DOI] [PubMed] [Google Scholar]
- Kussell, E., R. Kishony and N. Q. Balaban, 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169 1807–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann, M., and E. Jablonka, 1996. The inheritance of phenotypes: an adaptation to fluctuating environments. J. Theor. Biol. 181 1–9. [DOI] [PubMed] [Google Scholar]
- Leibler, S., and E. Kussell, 2010. Individual histories and selection in heterogeneous populations. Proc. Natl. Acad. Sci. USA 107 13183–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman, U., and M. W. Feldman, 1986. Modifiers of mutation rate: a general reduction principle. Theor. Popul. Biol. 30 125–142. [DOI] [PubMed] [Google Scholar]
- Lim, H. N., and A. van Oudenaarden, 2007. A multistep epigenetic switch enables the stable inheritance of DNA methylation states. Nat. Genet. 39 269–275. [DOI] [PubMed] [Google Scholar]
- Lynch, M., 2007. The Origins of Genome Architecture. Sinauer Associates, Sunderland, MA.
- Maynard Smith, J., 1978. The Evolution of Sex. Cambridge University Press, Cambridge, UK.
- Maynard Smith, J., 1980. Selection for recombination in a polygenic model. Genet. Res. 35 269–277. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., 1988. Selection for recombination in a polygenic model—the mechanism. Genet. Res. 51 59–63. [DOI] [PubMed] [Google Scholar]
- M'Gonigle, L. K., J. J. Shen and S. P. Otto, 2009. Mutating away from your enemies: the evolution of mutation rate in a host-parasite system. Theor. Popul. Biol. 75 301–311. [DOI] [PubMed] [Google Scholar]
- Nee, S., 1989. Antagonistic co-evolution and evolution of genotypic randomization. J. Theor. Biol. 140 499–518. [DOI] [PubMed] [Google Scholar]
- Rakyan, V. K., S. Chong, M. E. Champ, P. C. Cuthbert, H. D. Morgan et al., 2003. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc. Natl. Acad. Sci. USA 100 2538–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathé, M., J. Van Cleve and M. W. Feldman, 2009. Evolution of stochastic switching rates in asymmetric fitness landscapes. Genetics 182 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin, M., 2009. Epigenetic inheritance and the missing heritability problem. Genetics 182 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire, W., and G. Trapp, 1998. Using complex variables to estimate derivatives of real functions. SIAM Rev. 40 110–112. [Google Scholar]
- Thattai, M., and A. van Oudenaarden, 2004. Stochastic gene expression in fluctuating environments. Genetics 167 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland, R. A., and R. I. Jirtle, 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Biol. Evol. 23 5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngson, N. A., and E. Whitelaw, 2008. Transgenerational epigenetic effects. Annu. Rev. Genet. Hum. Genet. 9 233–257. [DOI] [PubMed] [Google Scholar]