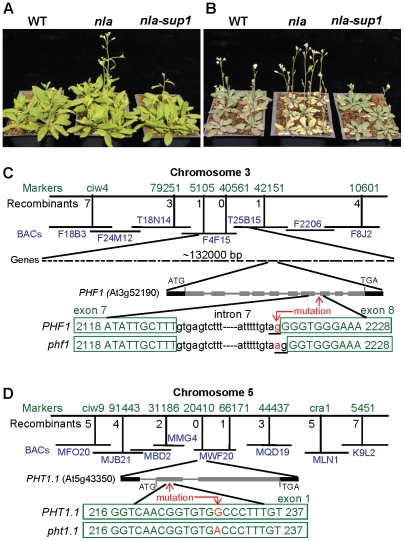
Figure 1. Recovery of the nla mutant phenotype by nla-suppressor1 and mapping of the nla-suppressors, PHF1 and PHT1.1.
(A) WT, nla and nla-sup1 (nla-phf1) plants grown at 10 mM NO3−-10 mM Pi. (B) Plants grown at 3 mM NO3−-10 mM Pi. (C) Mapping and partial genomic sequence of WT and nla-sup1 (phf1). The transition at 3′ end nucleotide ‘g’ to ‘a’, indicated by red arrow, disrupted the conserved dinucleotide, AG (underlined), which is required for splicing. In the phf1 mutation, the next available ‘G’ from the 5′ end of exon became part of the intron to form ‘AG’, causing loss of one G from the phf1 coding sequence. This cryptic splice site resulted in a frameshift and creation of a premature stop codon. (D) The mutation in the second nla suppressor was missense in PHT1.1 gene. In the first exon, nucleotide ‘G’ at position 229 was switched to ‘A’ which changes the codon GCC (encodes alanine) to ACC (for threonine).