Abstract
Antibodies microarrays are among the novel class of rapidly emerging proteomic technologies that will allow us to efficiently perform specific diagnosis and proteome analysis. Recombinant antibody fragments are especially suited for this approach but their stability is often a limiting factor. Camelids produce functional antibodies devoid of light chains (HCAbs) of which the single N-terminal domain is fully capable of antigen binding. When produced as an independent domain, these so-called single domain antibody fragments (sdAbs) have several advantages for biotechnological applications thanks to their unique properties of size (15 kDa), stability, solubility, and expression yield. These features should allow sdAbs to outperform other antibody formats in a number of applications, notably as capture molecule for antibody arrays. In this study, we have produced antibody microarrays using direct and oriented immobilization of sdAbs produced in crude bacterial lysates to generate proof-of-principle of a high-throughput compatible array design. Several sdAb immobilization strategies have been explored. Immobilization of in vivo biotinylated sdAbs by direct spotting of bacterial lysate on streptavidin and sandwich detection was developed to achieve high sensitivity and specificity, whereas immobilization of “multi-tagged” sdAbs via anti-tag antibodies and direct labeled sample detection strategy was optimized for the design of high-density antibody arrays for high-throughput proteomics and identification of potential biomarkers.
Keywords: Antibodies; metabolism; Antigens; immunology; Bacteria; metabolism; Biotinylation; Carcinoembryonic Antigen; immunology; Cell Extracts; Cell Line; Complex Mixtures; metabolism; Cost-Benefit Analysis; Cytoplasm; metabolism; Escherichia coli; metabolism; High-Throughput Screening Assays; economics; methods; Humans; Immobilized Proteins; metabolism; Immunoassay; Protein Array Analysis; economics; Tumor Markers, Biological; immunology
Keywords: Antibodies, single domain, cancer, diagnostic, array, beads
Introduction
The basic concept of microarray technology was initiated by the model of Ekins [1, 2] stating that “microspot” of elements should be able to detect analytes with a higher sensitivity than conventional immunoassays. On this basis, DNA microarray rapidly became the first application of this model and several well established approaches are now available to monitor mRNA abundance from small amount of materials. However, this approach is biased by the lack of relation between mRNA and proteins abundance, and is not sufficient to understand complex cellular networks. Considerable effort are being undertaken to develop a comparable technology for the analysis of protein levels, modifications and interactions. The development of methods for the global analysis of the protein of the cell is still in its early stage. Currently, the dominant technology for profiling of protein expression is two-dimensional (2D) gel electrophoresis [3]. This procedure is time-consuming and expensive and reproducibility is often an issue. There is a challenge to replace gels with alternative methods of protein expression analysis, including microarrays. Three types of protein microarrays are currently used: analytical, functional and reverse phase microarrays [4]. Analytical microarrays are typically used to profile a complex mixture of proteins in order to measure binding affinities, specificities and protein expression levels. In this technique, monoclonal antibodies or derived formats such as Fab (Fragment Antigen Binding), scFv (single chain variable Fragment) but also aptamers and affibodies [5] are arrayed on a support and the array is probed with a protein solution. Antibody microarrays, pioneered by MacBeath and Schreiber [6] and Haab et al [7], are the most common analytical microarray. This type of microarray will provide new means to perform differential protein expression profiling of healthy vs. diseased samples. They are expected to play a key role within disease diagnostics, biomarkers discovery and drug target identification. The ability to monitor multiple protein interactions in parallel has many advantages such as saving of time, cost, sample consumption, especially if assays are miniaturized. Most array-based strategies use sandwich assays that can be highly sensitive and specific, but this design is not compatible with high-density array. A complementary technology is label-based detection, affording high level of multiplexing and high density, at the expense of specificity and sensitivity [8].
To perform global proteome analysis, high demands will be placed upon the choice of catcher proteins. The specificity of the probes is also a critical feature since analytes must be specifically detected in heterogeneous mixtures containing more than 10 000 different irrelevant proteins. Currently, mainly low-density antibody microarrays (on planar substrate or on bead) have successfully been designed and developed [9–13]. In contrast to nucleic acids, antibodies and proteins in general are chemically and structurally much more complex, heterogeneous, and often unpredictable regarding their interaction profiles. Therefore, it is difficult to define general protein detection and immobilization strategies that do not discriminate between proteins. It is well accepted that oriented immobilization strategies improve array performances [14–16] but most described strategies involve multiple steps including purification and are not compatible with high-throughput array generation.
Recombinant antibody libraries such as scFv or Fab, providing numerous probes based on a single scaffold with similar biological properties, will display significant advantages. But, recombinant antibody formats such as scFv are often unstable [17] and produced with a poor yield. In 1993, Hamers-Casterman et al [18] discovered that serum of camels, dromedaries and llamas contain a unique type of antibodies devoid of light chains. Camelids produce functional antibodies devoid of light chains (HCAbs) and CH1 domain, of which the single N-terminal domain is fully capable of antigen binding. When they are recombinantly produced, these single domain antibody fragments (sdAbs) have several advantages for biotechnological applications thanks to their unique properties of size (15 kDa), stability even without disulfide bond formation [19], solubility, and expression yield [20]. These features should lead to a number of applications where sdAb should outperform other antibody formats, notably as capture molecule for antibody arrays.
In this study, we have generated proof-of-principle for several immobilization strategies of sdAbs contained in crude bacterial lysates, namely immobilization of in vivo biotinylated sdAbs by direct spotting of bacterial lysate on streptavidin, or multi-tagged sdAbs on anti-tag antibodies. By use of these immobilization strategies, we compared different detection methods, either by sandwich or label-based detection. These methods allow the specific and sensitive detection of subnanomolar antigen concentration without using signal amplification in model systems with pure antigen as well as crude patient sera.
Materials and methods
Proteins and serum sample
Anti-HIV-1 Nef sdAb (manuscript in preparation and [21]) and anti-CEA sdAb [22] were selected from immunized sdAb libraries. pET vector were used to produce in vivo biotinylated sdAbs. All sdAbs produced in this vector carry a C-terminal his6-tag, with or without Avitag™ (GLNDIFEAQKIEWHE) upstream. To generate plasmids coding for sdAb-avitag™ -his6, sdAb-tags was first amplified from pET-sdAbaNef-his6 using primers birA6hrev (TCAGCAAGCTTAGGATCCGTGATGATGATGGTGGTGTTCGTGCCATTCGATTTTCTGAGCCTCGAAGATGTCGTTCAGACCTGCGG CCGCTGAGGAGACAG) and seqT7 (TAATACGACTCACTATAGGG). Purified PCR product were digested with NcoI and BamHI and followed by gel purification and ligation into vector pET-sdAbaNef -his6 which had been previously digested with the same restriction enzymes.
pJF55 vector was used for the production of sdAbs fused to c-myc tag. All sdAbs produced in this vector contain one, three, or no C-terminal myc-tag (EQKLISEEDL) followed by a his6-tag. Vector pJF55-trimyc-his6 was generated by overlapping PCR using primers trimycfor (ACCGTCTCCTCAGCGGCCGCAGAACAGAAACTGATCTCTGAAGAGGACCTGAACGGTGAGCAGAAGCTCATTTCCGAGG) and trimycrev (CGCCAAAACAGAAGCTTTTAGTTGAGGTCCTCTTCGCTGATCAATTTTTGTTCGCCATTCAAATCTTCCTCGGAAATGAGCTTCTG C). Then, the purified PCR product was digested with NotI and HindIII, gel-purified and cloned into vector pJFsdAb-cmyc-his6 that had been digested with the corresponding restriction enzymes. All constructs were verified by nucleotide sequencing.
Patient sera were kindly provided by Pr. J.H. Cohen, (Université de Reims Champagne-Ardenne, Reims). Concentration of soluble CEA in patient sera varied between 150 and 750 ng/ml while CEA negative sera have a concentration of CEA lower than 5 ng/ml.
In vitro Biotinylation
The in vitro biotinylation of protein was performed using Ez-link micro NMHS-PEO4-biotinylation kit (Perbio science) following the recommendation of the manufacturer.
Labeling with Alexa488
The labeling of Nef with Alexa488 was performed using Alexa Fluor 488 Microscale Protein Labeling kit (Invitrogen) following the recommendation of the manufacturer to obtain a degree of labeling (DOL) of Nef of around 3 Alexa per molecule.
Production and purification of sdAbs
Vectors pET and pJF containing different sdAbs were transformed in Bl21DE3 and DH5 α strain respectively. Cells containing the plasmid were inoculated in 10 ml of 2YT medium (bactotryptone 16 g/l, yeast extract 10 g/l, NaCl 85 mM) supplemented with ampiciline (100 μg/ml) and glucose (2%). Cells were grown over night at 37 °C (250 rpm). Then cells were diluted to obtain an OD600 of 0.1 in 400 ml of 2YT medium supplemented with ampicillin (100 μg/ml) and cultures were grown until the OD600 reached 0.5, when sdAb expression was induced by the addition of 0.1 mM IPTG (isopropyl-h-D-thiogalactopyranoside) at 30°C (250 rpm) for 20 h. For in vivo biotinylated sdAbs, bacteria were co-transformed with pBir vector (Avidity, Colorado) and the culture medium was supplemented with chloramphenicol (50 μg/mL) during production. Fifty μM biotin was added during the induction.
Cells were harvested by centrifugation at 4000 rpm for 10 min at 4°C. For periplasmic purification, the cell pellet was suspended in 4 mL of cold TES buffer (0.2 m Tris ⁄ HCl, pH 8.0; 0.5 mM EDTA; 0.5 M sucrose), and 160 μL lysozyme (10 mg/mL) in TES buffer was added. Cells were subjected to osmotic shock by the addition of 16 mL of cold TES diluted 1/2 with cold H2O. After 30 min of incubation on ice, the suspension was centrifuged at 4000 rpm for 40 min at 4°C. The supernatant was incubated with 150 μL DNaseI (10 mg/mL) and MgCl2 (5 mM final) for 30 min at room temperature. The solution was dialyzed against 50 mM sodium acetate pH 7.0, 0.1 M NaCl, for 16 h at 4°C.
For cytoplasmic purification, cell pellet was frozen during 20 min at −80°C and lysed by 20 ml of bugbuster (Novagen) during 20 min with low shaking.
All sdAbs were purified by affinity chromatography on Talon™ metal affinity resin (Clontech). Bound molecules were eluted with 250 mM imidazole, and proteins were concentrated in PBS by ultrafiltration with Amicon Ultra 5000 MWCO (Millipore, Billerica, MA, USA) and stored at − 20°C. Their degree of purity was evaluated by SDS-PAGE analysis and protein concentration (average of 5 mg/ml) was determined spectrophotometrically using a protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA).
Production of sdAb-containing crude bacterial lysates
Vectors pET and pJF containing different sdAbs were transformed in BL21DE3 and DH5 α strain respectively. Transformed cells were inoculated in 96 well plates containing 150 μl/well of 2YT medium supplemented with ampicillin (100 μg/mL). Cells were grown until OD600 reached 0,5 and incubated 3 hours at 37°C after induction using 0.1 mM IPTG. For in vivo biotinylated sdAbs, bacteria were co-transformed with pBir vector and the culture medium was supplemented with chloramphenicol (50 μg/mL) during production and 50 μM of biotin was added during induction. After production, plates were centrifuged at 1700 rpm during 10 min and pellets were lysed with 30 μl of bugbuster during 20 min with low shaking. Plates were stored at − 20°C.
Cell lines
MC38-CEA and MC38 [23] are a kind gift of A. Pelegrin. Cells lines were cultured in DMEM complemented with 10% (v/v) fetal calf serum at 37°C in a humidified atmosphere and with 5% CO2. MC38-CEA culture medium was additionally complemented with 0.5 mg/ml of geneticin.
Flow cytometry analysis
Experiments were performed on ice with rocking in 1% BSA PBS. Typically, 2×105 cells resuspended in 50 μl were distributed in 96-well microtiter plate, and incubated for 1 h with various concentrations (500 to 0.00005 nM) of anti-CEA sdAb produced in the cytoplasm or periplasm of bacteria. After washing, binders were detected with anti-his6 mAb (Novagen) (1:1000). Washed cells were labeled with FITC conjugated anti-mouse antibody (Jackson) (1:60). Fluorescence was measured using a FACSCalicur™ (Becton and Dickinson) and results were analysed with the cellquest™ software. Negative (secondary antibody only) controls were carried out.
ELISA and slide assay
Activity of cytoplasmic and periplasmic sdAbs
Streptavidin plates (Thermo scientific) were blocked with 5% milk-PBS (MPBS) for two hours at RT. Fifty μl/well of biotinylated Nef at 5 nM in 2% MPBS were incubated overnight at 4°C. Wells were washed and incubated for 1h at RT with 50 μl of 2% MPBS containing various concentrations (500 to 0.00005 nM) of anti-Nef sdAb produced in the cytoplasm or periplasm of bacteria. After three washes with PBS, plates were incubated with 9E10 mAb (against c-myc) (santa cruz biotechnology) (1 μg/ml) in 2% MPBS for one hour at RT. Following three washes with PBS, a goat anti-mouse HRP-conjugated mAb (Jackson) (0.16 μg/ml in 2% MPBS) was incubated for one hour at RT. After three washes in PBS, bound secondary antibodies were detected using ABTS. Coloration was followed at 405 nm.
Immobilization of sdAbs biotinylated
Streptavidin plates (Thermo scientific), streptavidin beads (invitrogen), or nitrocellulose slide (Sciencetec) coated with streptavidin overnight at 4°C (10 μg/ml) were blocked with 5% MPBS for two hours at RT. SdAbs were diluted in 50 μL of 2% MPBS and incubated overnight at 4°C in streptavidin plate and in plate containing beads. SdAbs contain in bacterial lysate diluted ¼ in 2% MPBS were spotted and slides were dried for one hour at RT. Wells and slides were incubated with sample (Nef or serum) in 2% MPBS one hours at RT. After three washes with PBS, plates and slides were incubated with primary antibody (anti–Nef mouse mAb (kinf gift of Y. Collette, Marseille) 1:3000 or anti-CEA 35A7 antibody 2 μg/ml, (kind gift of A. Pelegrin, Montpellier) in 2% MPBS for one hour at RT. Following three washes with PBS, a goat anti-mouse HRP (Jackson) (0.16 μg/ml) or Alexa488-conjugated mAb for bead assay or Alexa680-conjugated mAb for slide assay (Invitrogen) (4 μg/ml) was incubated in 2% MPBS for one hour at RT. After three washes in PBS, plate with HRP labeled mAb was colorimetrically detected at 405 nm using ABTS substrate (Sigma), plate with Alexa labeled mAb was detected on Tristar reader (Berthold technologies) and slides with Alexa labeled mAb were read on Odyssey infrared imaging system (Licor).
Immobilization of trimyc-tagged sdAbs
Protein G beads or epoxy beads (Invitrogen), or nitrocellulose slides (Sciencetec) were incubated with 9E10 mAb (anti c-myc mAb, Santa-Cruz, sc-40) (1 μg/ml in PBS) 1h at RT for protein G beads, 48h at 4°C for epoxy beads, ON at 4°C for slides and blocked with 5% MPBS for two hours at RT. SdAbs, either pure or contained in crude bacterial lysate were diluted in 2% MPBS and incubated for 1h at RT in plate with containing beads. SdAbs contain in bacterial lysate diluted ¼ in 2% MPBS were spotted and slides were dry one hour at RT. After three washes with PBS, plates and slides were incubated with biotinylated Nef in 2% MPBS for one hours at RT. After 3 washes with PBS, plates were incubated with HRP-conjugated streptavidin (Jackson) (1 μg/ml) and slides with Alexa680-conjugated streptavidin (Invitrogen) (4 μg/ml). Following 3 washes in PBS, ABTS substrate (sigma) was added on plates and color development was followed at 405 nm and slides with Alexa labeled streptavidin were read on Odyssey infrared imaging system (Licor).
Results
1. Domain antibodies can be efficiently expressed in E. coli cytoplasm
Libraries of recombinant antibody fragments are a rich source of capture reagents. However, because they require disulfide bond formation, most fragments such as Fab or scFv fragments are produced in the periplasmic space of E. coli, an oxidizing environment favoring a correct folding of these fragments. In contrast, single domain antibodies are characterized by a very high solubility and stability that should allow them to fold properly in reducing environments such as the E. coli cytoplasm. To check this hypothesis, two model sdAbs (targeting Nef from HIV-1 [21] or human carcinoembryonic antigen (CEA) [22]) were produced in E. coli fused or not to a signal sequence, and purified from the periplasmic or cytoplasmic extract, respectively, and purified by metal affinity chromatography. As for most sdAbs, high production yields (10–30 mg.L−1) were obtained. Gel filtration analysis showed than only monomer format was produced (data not shown). Both versions of anti-CEA sdAbs were shown to perform similarly by flow cytometry on MC38-CEA cells, a murine colon carcinoma cell line transfected with human CEA cDNA [23] (Fig. 1A) and similar results were obtained with both versions of the anti-Nef sdAb by ELISA (Fig. 1B), demonstrating that sdAbs can be efficiently produced in an active form in the cytoplasm of E. coli.
Figure 1. Functional sdAbs are efficiently produced in the cytoplasm of E. coli.
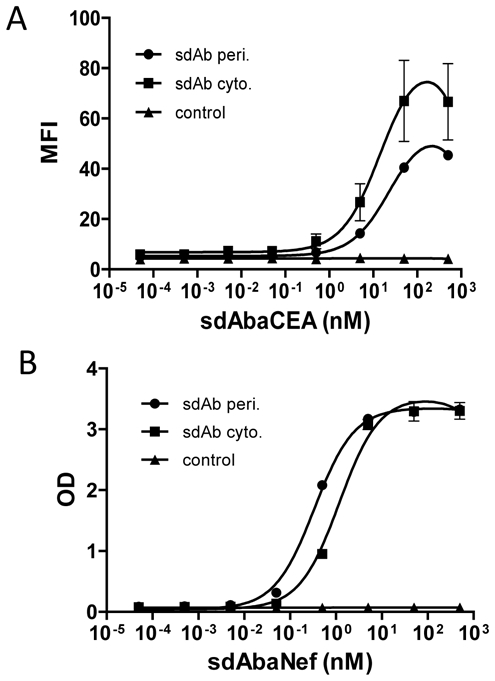
A) MC38-CEA (■, ●) or MC38 (▲) cells were incubated with serial dilutions of anti-CEA sdAb produced in cytoplasm (■) or periplasm (●) of E. coli. Captured antibodies were detected by a mouse anti-6his mAb followed by a goat against mouse FITC-conjugated mAb. Cells were analyzed by flow cytometry assay on FACScalibur. B) Biotinylated Nef antigen (5 nM) coated on streptavidin-plate was incubated with serial dilution of anti-Nef sdAb produced in the cytoplasm (■) or periplasm (●) of E. coli or anti-CEA sdAb produced in the cytoplasm of E. coli (▲). Captured antibodies were detected by a mouse anti c-myc mAb followed by a goat anti-mouse HRP-conjugated mAb. Standard deviation represents two experiments performed in triplicates.
2. Oriented sdAb immobilization
Beside its efficiency, cytoplasmic sdAb production further offers the possibility to biotinylate sdAbs in vivo using a C-terminal fusion with a 15 amino acids tag (avitag) recognized by the E coli BirA enzyme. The resulting molecules possess a single biotin molecule coupled to a single lysine present on the avitag, which allows a near covalent and oriented immobilization through binding to streptavidin. In contrast, in vitro biotinylation can lead to inactivation of the protein and does not allow oriented immobilization. To test this hypothesis, the anti-Nef sdAb was fused to the avitag, biotinylated in vivo and purified. For comparison, the anti-Nef sdAb was purified and biotinylated in vitro using a primary amine coupling strategy. Biotinylation efficiency was checked by incubation over streptavidin beads. Up to 95% of in vivo biotinylated sdAbs and 80% of in vitro biotinylated sdAbs could be captured on beads, demonstrating an efficient biotinylation (data not shown). As shown in Fig. 2A, the in vivo biotinylated sdAb had to be diluted tenfold compared to the chemically biotinylated version to yield similar results, suggesting that in vivo biotinylation preserves the activity of the sdAb and allows an optimal orientation.
Figure 2. In vivo biotinylation and multi tags strongly improve immobilization of sdAbs.
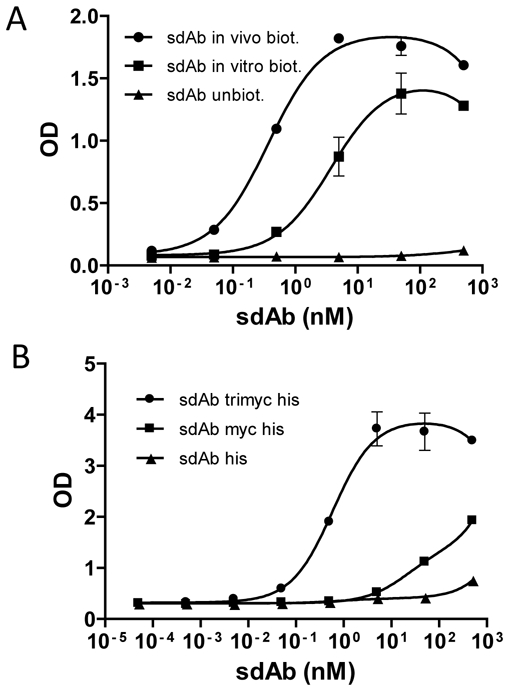
A) Serial dilutions of pure anti-Nef sdAb biotinylated in vivo (●), in vitro (■) or unbiotinylated (▲) were coated on streptavidin plate and incubated with Nef at 5 nM. The captured antigen was detected with a mouse anti-Nef antibody followed by a goat anti-mouse HRP-conjugated mAb. B) Protein G bead were coated with 1 μg/ml of 9E10 mAb. Serial dilutions of pure sdAbs with three (●), one (■) or no (▲) myc tag were incubated with the bead, followed by biotinylated Nef at 5 nM. The captured antigen was detected with a HRP-conjugated streptavidin. Standard deviation represents two experiments performed in triplicates.
However, this strategy precludes the use of the streptavidin:biotin system for detection of bound molecules, a useful strategy for high throughput approaches. As an alternative, we decided to immobilize sdAbs via interaction between a c-terminal c-myc tag (EQKLISEEDLN) and mAb 9E10, a commercially available murine IgG binding to this peptidic sequence. Three tandem repetitions of the tag (named trimyc tag) were also fused to the sdAb C-terminus to increase the apparent affinity of the tagged molecule by avidity effect. As demonstrated by ELISA, (Fig. 2B), mAb 9E10 bound to protein G beads led to a much better sdAb immobilization, leading in turn to a much higher capture efficiency of the model antigen. The sdAb bearing the trimyc tag could be diluted by three orders of magnitude compared to the molecule bearing a single c-myc tag to yield similar signals.
3. Use of crude lysates containing sdAbs
The highly efficiencies reached by these immobilization strategies allow the use of very low concentration of capture sdAbs. We reasoned that the oriented immobilization could be used as a built-in purification procedure, allowing the use of crude bacterial lysates. Indeed as demonstrated in Fig. 3, as low as 50 nL of a crude lysate containing the in vivo biotinylated anti-Nef sdAb yielded the same signal intensity as 1 μg/mL of the same purified sdAb on a bead assay, suggesting that a regular microplate sdAb production (30 μL) could be used to generate up to 600 measures.
Figure 3. Bacterial lysates are a good source of capture antibody.
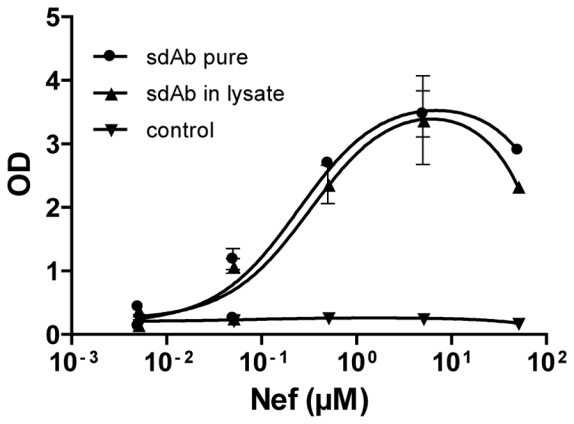
Streptavidin beads were coated with sdAb against Nef biotinylated in vivo pure (●) (1 μg/ml) or in bacterial lysate (▲) (50 nl/wells) or sdAb against Nef unbiotinylated in bacterial lysate (▼) and incubated with serial dilution of Nef. The captured antigen was detected with a mouse anti-Nef antibody followed by a goat anti-mouse HRP-conjugated mAb. Standard deviation represents two experiments performed in triplicates.
4. Assay Sensitivity
To determine the sensitivity of the trimyc-based assay, beads coupled to 9E10 were incubated with 50 nL of bacterial lysate containing the anti-Nef sdAb fused to the c-myc or the trimyc tag. Beads were subsequently incubated with various concentrations of biotinylated Nef. Bound antigen was detected using streptavidin-HRP. Fig. 4A shows that the trimyc tag yielded much higher signals, leading to the detection of 0.5 nM of Nef. Similar results were obtained using nitrocellulose arrays. Moreover, 9E10 incubation of slides led to a significant decrease of background noise (Fig. 4B). Interestingly, the single c-myc tag yielded much lower signals. Of note, a direct labeling of Nef with a fluorescent probe (Alexa 488) yielded very poor results in both settings, despite a normal affinity between the Alexa 488-Nef and anti-Nef sdAb (data not shown).
Figure 4. In vivo biotinylated and trimyc-tagged sdAbs in bacterial lysate allow a sensitive antigenic detection on slide or beads.
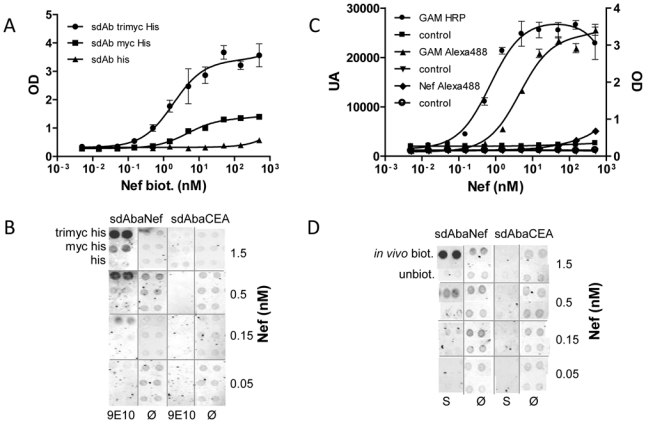
A) Immobilization of sdAbs using trimyc tag allows the direct use of bacterial lysates. Epoxy bead were coated with 9E10 (1 μg/ml) and incubated with bacterial lysate (50 nl/wells) containing sdAbs with three (●), one (■) or no (▲) c-myc followed by serial dilutions of biotinylated Nef. The captured antigen was detected with HRP-conjugated streptavidin. B) Immobilization of sdAbs using trimyc allows direct spotting of bacterial lysate. Nitrocellulose slides were coated with 9E10 (9E10) or PBS (Ø). Bacterial lysates containing sdAbs fused to three, one or no c-myc were spotted. Serial dilutions of biotinylated Nef were incubated and the captured antigen was detected with Alexa705-conjugated streptavidin. C): Streptavidin beads were coated with bacterial lysate (50 nl/wells) containing anti-Nef sdAbs biotinylated in vivo (●, ▲, ◆) or not (■, ▼, ○) and incubated with serial dilutions of Nef or Alexa488-conjugated Nef (◆, ○). The captured antigen was detected with a mouse anti-Nef antibody followed by a goat anti-mouse HRP (●,■) or Alexa488-conjugated mAb (▲, ▼). D) Nitrocellulose slides were incubated with streptavidin (S) or PBS (Ø). Bacterial lysates containing sdAbs biotinylated in vivo or unbiotinylated were spotted. Serial dilutions of Nef were incubated and the captured antigen was detected with a mouse anti-Nef antibody followed by a goat anti-mouse Alexa705-conjugated mAb. Standard deviation represents two experiments performed in triplicates.
Similar experiments were conducted using the streptavidin/avidin based immobilization strategy and 50 nL of crude bacterial lysate per well. Detection of the bound antigen was performed using three different methods, namely using an anti-Nef mAb followed by a HPR-labeled secondary antibody or an Alexa-labeled secondary antibody, compared to a direct fluorescent labeling of the antigen (Nef-Alexa). As shown in Fig. 4C, the indirect labeling strategies yielded the best results, with a detection limit of 0.5 nM for enzymatic indirect labeling, 5 nM for fluorescent indirect labeling and 50 nM for the fluorescent direct labeling. A detection limit of 0.5 nM was measured on nitrocellulose slides using sandwich fluorescent detection. Streptavidin preincubation of slides further led to a significant decrease of background noise (Fig. 4D).
5. Application to clinically relevant concentration of cancer biomarker
To demonstrate that these strategies can be applied to high throughput diagnostic approaches, 0.5 μL of crude bacterial lysates containing in vivo biotinylated anti-CEA sdAbs were used in a bead based assay to detect soluble CEA in serial dilutions of crude cancer patient sera of known CEA concentration. Detection was performed using an enzymatic sandwich assay. As shown in Fig. 5, soluble CEA could be detected in all CEA-containing sera. The detection limit was established at 10 pM i.e. 2 ng/mL of soluble CEA. This concentration is below the value of circulating CEA in sera of normal donors (~ 5 ng/mL in undiluted serum) and well below concentration of cancer patients (178–769 ng/mL).
Figure 5. Immobilization of in vivo biotinylated sdAbs allows a sensitive detection of CEA in patient sera using a bead assay.
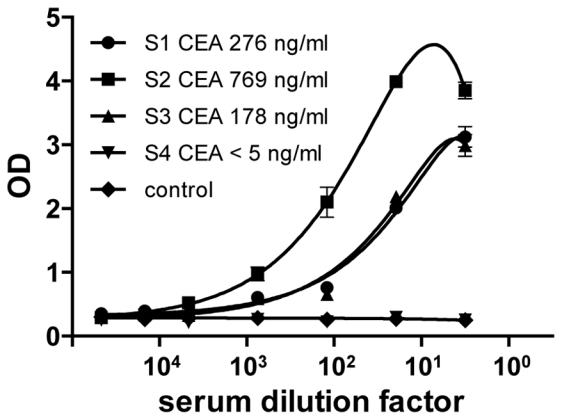
Streptavidin beads were coated with bacterial lysates (0.5 μl/wells) containing in vivo biotinylated (●,■, ▼, ▲) or unbiotinylated (◆) anti-CEA sdAbs and incubated with serial dilutions of patient sera (●: S1 CEA 276 ng/ml, ■: S2 CEA 769 ng/ml, ▲: S3 CEA 178 ng/ml, ▼: S4 CEA < 5ng/ml). The captured antigen was detected with a mouse anti-CEA antibody (35A7) followed by a goat anti-mouse HRP-conjugated mAb. Standard deviation represents two experiments performed in triplicates.
Discussion
Most antibody arrays developed to date are low density arrays relying on the use of pure preparation of intact monoclonal antibodies [24]. The requisite for high concentration of pure proteins is hindering the development of high density antibody arrays (in the 200–2000 μg/ml range). Recombinant antibodies such as scFv fragment offer an interesting alternative since this format is compatible with the generation of scFv libraries and high throughput selection methods such as phage or ribosome display. Unfortunately, those fragments are constituted by the association of two domains (VH and VL) which decreases their stability. Consequently, very high concentration of pure fragments (around 400 μg/mL) are often used to build microarrays [10, 25], which severely complicate the building process of high density antibody arrays.
In this study, we show that highly functional and sensitive arrays could be generated using non-purified affinity tagged single domain antibodies (sdAbs) as probes. These fragments are very easy to produce in E. coli, are compatible with cytoplasmic expression and are extremely stable. sdAbs were produced in 96 well plate format and successfully coupled, enriched and purified in a one-step procedure directly onto the support. Indeed, we demonstrate that extremely low amount, i.e. 0.5 to 0.05 μl (probably depending on the sdAb affinity) of crude bacterial lysate produced in three hours is sufficient to perform one assay. Such efficiency was achieved using strong and oriented immobilization on slide arrays or beads, through the use of directed cytoplasmic biotinylation of sdAbs for immobilization on streptavidin coated supports or a tandem repetition of the c-myc tag for immobilization on mAb 9E10-coated support. Oriented immobilization based on modified with Ni2+-ions [14] or streptavidin [15, 16, 26] are examples of surface that have been successfully applied to generate planar protein arrays through specific coupling chemistries. However, to our knowledge, only purified monoclonal antibodies coupled with standard procedure such as carbodiimide and succinimide reactions are currently used for bead arrays.
These two complementary approaches were developed to fulfill two different needs. One hand, high sensitivity and specificity are two crucial parameters for diagnostic arrays. In this case, the most efficient approach is the sandwich assay, using a pair of probes to specifically capture and detect the antigen of interest. In this case, non-purified sdAbs can be efficiently immobilized using the biotin/streptavidin setting, to be used as capturing reagent. This method allowed a subnanomolar limit of detection (LOD) of a pure model antigen Nef using fluorescent and enzymatic detection methods. In a clinical setting, i.e. the detection of circulating CEA in sera of cancer patients, a picomolar LOD of CEA in crude serum was obtained with an enzymatic sandwich detection system. In the case of Nef detection, slides or beads as assay support yielded similar results. Of note, direct labeling of antigen with fluorophore was found very inefficient, and chemical sample biotinylation followed by detection with labeled streptavidin led to much higher signals, as already demonstrated by other studies [27]. Bead assays are especially suited for sandwich assays and can be directly compared to ELISA method [28], while requiring much smaller volumes of sample material. Beads can be coded by using various concentrations of fluorescent dye, or by some type of barcoding technology such as size of the bead. Consequently, bead assays can easily be multiplexed. Thus bead arrays are method of choice for low density antibodies array for clinical diagnosis [29, 30]. In this work we show that magnetic beads can efficiently be functionalized using a biotin-based sdAb immobilization. This approach would therefore be the method of choice for the development of cost-efficient sandwich-based antibody bead arrays for diagnostic.
On the other hand, and despite its efficiency, the sandwich assay, requiring a pair of specific probe for each antigen, is not compatible with the generation of high density antibody arrays. These approaches most often necessitate sample labeling (i.e serum or cell lysate), being most efficiently performed using chemical biotinylation and labeled streptavidin [27]. For such needs, trimyc-tagged sdAbs were developed to similarly achieve strong and oriented immobilization without involving biotin. Very large sdAb libraries can be built in a one step procedure from immunized llamas without being concerned about correct combination of VH and VL domain, and rapidly enriched for specific binder by display techniques. Moreover, sdAbs are very efficiently produced in 96 well plates and thus, they represent a rich source of probe to generate high density arrays in a high throughput fashion. In this work, we demonstrate that high sensitivities in the nanomolar range could be achieved with this setting for our model antigen on beads but also on planar arrays such as nitrocellulose arrays, clearly more adapted to high density arrays, and demonstrating the feasibility of using crude bacterial lysate to immobilize tagged sdAbs on slide in a high throughput screening compatible fashion. This approach can further be used for differential screening (i.e. using normal vs disease samples) of sdAb libraries enriched on disease material, potentially leading to the discovery of new biomarkers. We are currently applying this approach to isolate breast cancer specific sdAbs from libraries built using animals immunized with breast cancer biopsies.
Supplementary Material
Acknowledgments
This work was supported by CNRS, INSERM and by the French National Research Agency (Agence Nationale de Recherche – ANR) program ‘Nanosciences and Nanotechnologies’ under the grant ANR-07-PNANO-051-01. We would like to thank J.H. Cohen and B. Reveil for the kind gift of patient sera.
Abbreviations
- mAb
monoclonal antibodies
- scFv
single chain Fv fragment
- sdAb
single domain antibodies
References
- 1.Ekins R, Chu F, Biggart E. Multispot, multianalyte, immunoassay. Ann Biol Clin (Paris) 1990;48:655–666. [PubMed] [Google Scholar]
- 2.Ekins R, Chu FW. Microarrays: their origins and applications. Trends Biotechnol. 1999;17:217–218. doi: 10.1016/s0167-7799(99)01329-3. [DOI] [PubMed] [Google Scholar]
- 3.Marcus K, Joppich C, May C, Pfeiffer K, et al. High-resolution 2DE. Methods Mol Biol. 2009;519:221–240. doi: 10.1007/978-1-59745-281-6_14. [DOI] [PubMed] [Google Scholar]
- 4.Joos T, Bachmann J. Protein microarrays: potentials and limitations. Front Biosci. 2009;14:4376–4385. doi: 10.2741/3534. [DOI] [PubMed] [Google Scholar]
- 5.Renberg B, Nordin J, Merca A, Uhlen M, et al. Affibody molecules in protein capture microarrays: evaluation of multidomain ligands and different detection formats. J Proteome Res. 2007;6:171–179. doi: 10.1021/pr060316r. [DOI] [PubMed] [Google Scholar]
- 6.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 7.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2:RESEARCH0004. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rousserie G, Sukhanova A, Even-Desrumeaux K, Fleury F, et al. Semiconductor quantum dots for multiplexed bio-detection on solid-state microarrays. Crit Rev Oncol Hematol. 74:1–15. doi: 10.1016/j.critrevonc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Miller JC, Zhou H, Kwekel J, Cavallo R, et al. Antibody microarray profiling of human prostate cancer sera: antibody screening and identification of potential biomarkers. Proteomics. 2003;3:56–63. doi: 10.1002/pmic.200390009. [DOI] [PubMed] [Google Scholar]
- 10.Carlsson A, Wingren C, Ingvarsson J, Ellmark P, et al. Serum proteome profiling of metastatic breast cancer using recombinant antibody microarrays. Eur J Cancer. 2008;44:472–480. doi: 10.1016/j.ejca.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 11.Ingvarsson J, Wingren C, Carlsson A, Ellmark P, et al. Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics. 2008;8:2211–2219. doi: 10.1002/pmic.200701167. [DOI] [PubMed] [Google Scholar]
- 12.Sauer G, Schneiderhan-Marra N, Kazmaier C, Hutzel K, et al. Prediction of nodal involvement in breast cancer based on multiparametric protein analyses from preoperative core needle biopsies of the primary lesion. Clin Cancer Res. 2008;14:3345–3353. doi: 10.1158/1078-0432.CCR-07-4802. [DOI] [PubMed] [Google Scholar]
- 13.Lyon DE, McCain NL, Walter J, Schubert C. Cytokine comparisons between women with breast cancer and women with a negative breast biopsy. Nurs Res. 2008;57:51–58. doi: 10.1097/01.NNR.0000280655.58266.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H, Bilgin M, Bangham R, Hall D, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 15.Peluso P, Wilson DS, Do D, Tran H, et al. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 16.Pavlickova P, Knappik A, Kambhampati D, Ortigao F, Hug H. Microarray of recombinant antibodies using a streptavidin sensor surface self-assembled onto a gold layer. Biotechniques. 2003;34:124–130. doi: 10.2144/03341rr03. [DOI] [PubMed] [Google Scholar]
- 17.Honegger A. Engineering antibodies for stability and efficient folding. Handb Exp Pharmacol. 2008:47–68. doi: 10.1007/978-3-540-73259-4_3. [DOI] [PubMed] [Google Scholar]
- 18.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 19.Gueorguieva D, Li S, Walsh N, Mukerji A, et al. Identification of single-domain, Bax-specific intrabodies that confer resistance to mammalian cells against oxidative-stress-induced apoptosis. FASEB J. 2006;20:2636–2638. doi: 10.1096/fj.06-6306fje. [DOI] [PubMed] [Google Scholar]
- 20.Muyldermans S. Single domain camel antibodies: current status. J Biotechnol. 2001;74:277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 21.Baty D, Chartier M, Chames P, Benichou S, et al. WO/2009/066241. 2009
- 22.Behar G, Chames P, Teulon I, Cornillon A, et al. Llama single-domain antibodies directed against nonconventional epitopes of tumor-associated carcinoembryonic antigen absent from nonspecific cross-reacting antigen. FEBS J. 2009;276:3881–3893. doi: 10.1111/j.1742-4658.2009.07101.x. [DOI] [PubMed] [Google Scholar]
- 23.Clarke P, Mann J, Simpson JF, Rickard-Dickson K, Primus FJ. Mice transgenic for human carcinoembryonic antigen as a model for immunotherapy. Cancer Res. 1998;58:1469–1477. [PubMed] [Google Scholar]
- 24.Knezevic V, Leethanakul C, Bichsel VE, Worth JM, et al. Proteomic profiling of the cancer microenvironment by antibody arrays. Proteomics. 2001;1:1271–1278. doi: 10.1002/1615-9861(200110)1:10<1271::AID-PROT1271>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Orchekowski R, Hamelinck D, Li L, Gliwa E, et al. Antibody microarray profiling reveals individual and combined serum proteins associated with pancreatic cancer. Cancer Res. 2005;65:11193–11202. doi: 10.1158/0008-5472.CAN-05-1436. [DOI] [PubMed] [Google Scholar]
- 26.Andresen H, Bier FF. Peptide microarrays for serum antibody diagnostics. Methods Mol Biol. 2009;509:123–134. doi: 10.1007/978-1-59745-372-1_8. [DOI] [PubMed] [Google Scholar]
- 27.Zajac A, Song D, Qian W, Zhukov T. Protein microarrays and quantum dot probes for early cancer detection. Colloids Surf B Biointerfaces. 2007;58:309–314. doi: 10.1016/j.colsurfb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Elshal MF, McCoy JP. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods. 2006;38:317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwenk JM, Gry M, Rimini R, Uhlen M, Nilsson P. Antibody suspension bead arrays within serum proteomics. J Proteome Res. 2008;7:3168–3179. doi: 10.1021/pr700890b. [DOI] [PubMed] [Google Scholar]
- 30.Rimini R, Schwenk JM, Sundberg M, Sjoberg R, et al. Validation of serum protein profiles by a dual antibody array approach. J Proteomics. 2009;73:252–266. doi: 10.1016/j.jprot.2009.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


