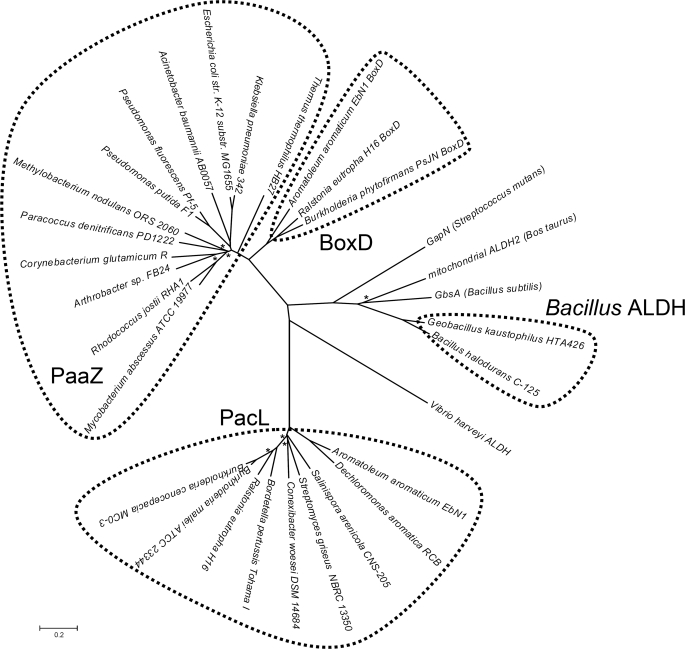
FIGURE 6.
Phylogenetic unrooted tree of a selection of orthologues of PaaZ-ALDH, PacL, Bacillus ALDH, BoxD, and some ALDHs that are not involved in aromatic degradation. The tree was constructed using the Neighbor-Joining method (28) and is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. Asterisks indicate bootstrap values <70%. ALDHs that are not involved in aromatic catabolism are: GapN, glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans (64) (accession number Q59931); mitochondrial bovine ALDH2 (53) (accession number P20000); GbsA: glycine betaine aldehyde dehydrogenase from B. subtilis (30) (accession number NP_390984); Vibrio harveyi ALDH (60) (accession number AAA89078).