Abstract
We have incorporated, for the first time, FtsZ and FtsA (the soluble proto-ring proteins from Escherichia coli) into bacterial giant unilamellar inner membrane vesicles (GUIMVs). Inside the vesicles, the structural organization and spatial distribution of fluorescently labeled FtsZ and FtsA were determined by confocal microscopy. We found that, in the presence of GDP, FtsZ was homogeneously distributed in the lumen of the vesicle. In the presence of GTP analogs, FtsZ assembled inside the GUIMVs, forming a web of dense spots and fibers. Whereas isolated FtsA was found adsorbed to the inner face of GUIMVs, the addition of FtsZ together with GTP analogs resulted in its dislodgement and its association with the FtsZ fibers in the lumen, suggesting that the FtsA-membrane interaction can be modulated by FtsZ polymers. The use of this novel in vitro system to probe interactions between divisome components will help to determine the biological implications of these findings.
Keywords: Cytoskeleton, Molecular Motors, Protein Assembly, Protein Self-assembly, Protein-Protein Interactions, FtsZ, Bacterial Cell Division
Introduction
Toward the end of the cell cycle, the bacterial cell division machinery assembles, forming a ring at mid-cell (1–3). This ring is a highly dynamic structure composed of at least 15 proteins, most of them integral membrane proteins. In Escherichia coli, the elements of the ring follow an assembly pathway in which both sequential and concerted stages are involved. Initially, the concerted action of FtsZ, FtsA, and ZipA results in the assembly of a proto-ring. The remaining proteins are incorporated later into a multiprotein complex that spans the cell membrane and is attached to the peptidoglycan. This complex directs the synthesis and eventually the modification of the peptidoglycan at mid-cell, leading to septation (3–5). Among the proto-ring elements, FtsZ, a GTPase similar to tubulin in structure but different in sequence, is probably the most ubiquitous prokaryotic division protein (6). FtsZ is a molecular machine capable of assembling into single-stranded filaments that further associate among themselves (7–12). FtsZ interacts with other division ring components, particularly the membrane-anchoring proteins FtsA and ZipA (3). FtsA is a member of the actin family with a short amphipathic helix that seems to mediate its association with the membrane (13–15). ZipA has an N-terminal helix that is integrated into the membrane and connected to a cytoplasmic FtsZ-interacting domain via a flexible linker (16). Apparently either FtsA or ZipA can support the attachment of FtsZ to the membrane, but if both are absent, FtsZ does not localize at the membrane (17).
In this work, we describe a procedure to obtain bacterial giant unilamellar inner membrane vesicles (GUIMVs)4 and its application to study the assembly and interactions of two proto-ring components, namely FtsZ and FtsA. These bacterial membrane vesicles were formed at physiological ionic strength, and being more complex than artificial lipid vesicles, they maintained a composition of proteins and lipids resembling more closely the native membranes. To mimic the crowded bacterial interior, the entrapment of the proto-ring proteins was carried out in the presence of high concentrations of inert macromolecules. The structural organization and spatial distribution of incorporated fluorescently labeled FtsZ and FtsA (either as isolated species or both simultaneously) in the lumen of GUIMVs were then studied by confocal microscopy.
EXPERIMENTAL PROCEDURES
Materials
Reagents, salts, protease inhibitors, and buffer components were from Sigma and Thermo Scientific. The GTP analogs P3-1-(2-nitro)phenylethyl-caged GTP and GMPPCP (a non-hydrolyzable form of GTP) were from Jena Bioscience. Protein A-HRP conjugate was purchased from Bio-Rad. The chemiluminescence detection system and Ficoll (average Mr of 70,000) were from GE Healthcare. Fluorescent dyes (Alexa 488, Alexa 647, and DiIC18 (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine)) were from Molecular Probes.
Proteins
E. coli FtsZ was purified as described previously (18). The protein was equilibrated in 50 mm Hepes/HCl (pH 7.4), 500 mm KCl, and 10 mm MgCl2 in the presence of 1 mm GDP. FtsA was produced from E. coli strain BL21(DE3) harboring pMFV12 expressing an N-terminal fusion of ftsA+ to a His tag (19) and purified according to a method optimized recently in our laboratories that allows FtsA to be obtained from inclusion bodies.5 The refolded protein is stable in solution (with the main species having an s value of 3 S) and retains the ability to interact with FtsZ (see “Results”).
Protein Labeling
FtsZ (4 mg/ml) and FtsA (0.4 g/liter) were labeled with Alexa probes (1:10 molar ratio) in 50 mm Hepes/HCl (pH 8.0), 100 mm KCl, and 5 mm MgCl2 at room temperature for 30 min. FtsZ was labeled under conditions that promote protein polymerization to ensure minimal interference of the dye with FtsZ assembly as described by González et al. (21). Labeled proteins were separated from free probe using a gel filtration column, distributed in aliquots, frozen in liquid nitrogen, and stored at −80 °C. The degree of labeling was 0.9 ± 0.2 mol of fluorophore/mol of protein. There was no difference in the behavior of both labeled proteins compared with the unlabeled proteins. For example, fluorescently labeled FtsZ had the same critical concentration for assembly and response to solution conditions to polymerize as did WT FtsZ.6 Alexa 488 and Alexa 647 were chosen to avoid fluorescence transfer.
Isolation of E. coli Inner Membranes
Inner membrane vesicles were isolated from wild-type E. coli (strain JM600) exponential phase culture (20) essentially as described by De Vrije et al. (22). The inner and outer membrane vesicles were separated by sucrose gradient centrifugation according to Osborn et al. (23), washed and diluted to reach 20 absorbance units at 280 nm, and stored frozen at −80 °C.
Giant Unilamellar Vesicle (GUV) Preparation from E. coli Inner Membranes
GUIMVs were prepared by electroformation under physiological salt conditions as described by Pott et al. (24) using a homemade chamber with platinum electrodes (25, 26). Aliquots of inner membrane vesicles (4 μl) were seeded on each platinum electrode at 37 °C. Preheated reconstitution buffer (50 mm Tris-HCl (pH 7.4), 100 mm KCl, 100 mm sucrose, and 50 mg/ml Ficoll 70) was added to the samples.
Reconstitution of Proto-ring Elements inside GUIMVs
Where indicated, FtsZ and FtsA (fluorescently labeled or not) and the corresponding nucleotide were added to the chamber to incorporate these division proteins inside the vesicles. Most of the experiments were done with FtsZ/FtsA mixtures at the concentrations given by Rueda et al. (20), namely 5 and 1 μm, respectively. Similar results were obtained with concentrations of 10 and 2 μm, respectively. The localization of ZipA and FtsN on GUIMVs was performed as described by Montes et al. (25) with anti-ZipA antibody MVC1 (1:1000) (20), anti-FtsN antibody MVG1 (1:1000) (27), and Alexa 488-labeled anti-rabbit IgG.
To obtain stable FtsZ polymers during the time scale of the experiments (∼2 h), protein assembly was triggered upon addition of 5 mm MgCl2 and 0.5 mm GTP analog in the presence of 50 mg/ml Ficoll (a crowding agent that promotes FtsZ assembly to form ribbons and bundles (21)). GUIMVS were also formed in the absence of Ficoll, but as expected, FtsZ assembled into protofilament fibers that were too narrow to be visualized by confocal microscopy. The figures shown in this work correspond to FtsZ polymers formed in the presence of caged GTP, but the same results were obtained with GMPPCP (data not shown). GUIMVs were directly observed by confocal microscopy using a Leica TCS SP5 microscope with an Acousto optical beam splitter and a 100× (1.4–0.7 numerical aperture) oil immersion objective. The excitation wavelengths were 633, 533, and 488 nm (for Alexa 647, DiIC18, and Alexa 488, respectively). When caged GTP was used to trigger FtsZ assembly, GUIMV formation was carried out in the dark, and the photolysis of the caged nucleotide was induced at 350 nm by a UV laser. Image processing was performed using NIH ImageJ (rsb.info.nih.gov/ij/).
Assay of FtsA Binding to Inner Membranes
Inner membrane vesicle fractions (100 μl at 1 mg/ml) were incubated with Alexa 488-labeled FtsA (1 μm final concentration) in 50 mm Tris-HCl and 100 mm KCl (pH 7.4) for 30 min at room temperature and centrifuged at 13,000 rpm for 10 min. To remove free FtsA, the resulting membrane pellet was extensively washed and centrifuged until the protein signal was undetectable/negligible in the supernatant. Unlabeled FtsZ (25 μm), MgCl2 (10 mm), and GTP/ATP (1 mm) were added, and the FtsZ-FtsA heteropolymers were detected in the supernatant. In each step, the presence of both proteins was assayed by SDS-PAGE followed by Western blotting with anti-FtsZ antibody MVJ9 (28) and anti-FtsA antibody MVM1 (14) using standard protocols (29). The antibodies were detected with protein A coupled to peroxidase using chemiluminescence.
RESULTS
Production of Bacterial GUIMVs
Giant vesicles made exclusively from the bacterial inner membrane were formed under physiologically relevant ionic strength conditions (100 mm KCl) and in the presence of high concentrations of inert macromolecules (50 mg/ml Ficoll 70) to mimic the crowded bacterial interior (30, 31). Both multi- and unilamellar vesicles were observed ranging in size from 5 to 50 μm. Only unilamellar vesicles were analyzed in this work. A typical GUIMV, in which the vesicle unilamellar structure was visualized using the lipid dye DiIC18, is shown in Fig. 1 (left panel). The differential interference contrast image is shown in supplemental Fig. S1. To characterize these vesicles initially, the orientation of relevant membrane proteins was determined. For this purpose, two membrane proteins were selected and identified using immunofluorescence detection: ZipA, the proto-ring protein that anchors FtsZ at the cytoplasmic side of the membrane, and FtsN, a cell division protein that faces the periplasmic lumen. Fig. 1 (middle and right panels) shows GUIMVs labeled with specific anti-ZipA and anti-FtsN fluorescent markers. GUIMVs were visualized with the two markers, suggesting that ZipA is located on both sides of the vesicles. These results indicate that the protocol used to isolate the inner membranes yields a mixed population of inverted and right-side-out membranes at a ratio of ∼6:4 (22, 32).
FIGURE 1.
Image analysis of GUVs obtained from E. coli inner membranes. Left panel, single equatorial confocal image of a GUIMV stained with the lipid dye DiIC18 (DiI). Scale bar = 30 μm. Middle and right panels, immunolocalization of ZipA and FtsN in GUIMVs. Shown are single equatorial images of a GUIMV in the presence of anti-ZipA (middle panel) or anti-FtsN (right panel) antibody (see “Experimental Procedures”). Scale bars = 10 μm.
FtsZ Can Be Incorporated and Visualized inside GUIMVs as a Web of Dense Spots and Fibers
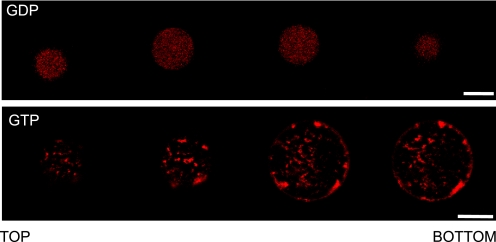
During GUIMV formation, FtsZ in its GDP-bound state was trapped inside the vesicles (Fig. 2, upper panel). For the sake of brevity, this figure shows alternative sections, which correspond to the complete series included in supplemental Fig. S2. To visualize the protein under a confocal microscope, a 1:10 molar ratio of FtsZ labeled with Alexa 647 was used (see “Experimental Procedures”). The complete series of cross-sectional images from these preparations showed that most of GDP-FtsZ remained soluble and homogeneously distributed inside the vesicles, with few detectable interactions with the inner surface. In the presence of a GTP analog (with photolysis of the caged nucleotide induced by a UV laser), FtsZ polymerized, and the spatial distribution of the protein inside the vesicle was modified (Fig. 2, lower panel, and supplemental Fig. S3). In this case, FtsZ was not distributed homogeneously but localized in dense structures both at the inner surface of the vesicle and throughout the lumen, which is better appreciated when individual sections are visualized. In this sequence, some spots of FtsZ in the proximity of the membrane and additional dispersed fibers inside the vesicle were observed.
FIGURE 2.
Spatial distribution of Alexa 647-labeled FtsZ inside GUIMVs. Upper panel, image analysis of FtsZ inside GUIMVs formed in the presence of 0.1 mm GDP. Shown are sequential cross-sectional images (taken every 4 μm) of GDP-FtsZ (Alexa 647-labeled) inside GUIMVs. The whole collection of images showed the same pattern (supplemental Fig. S2C). Lower panel, image analysis of FtsZ inside GUIMVs formed in the presence of 0.5 mm caged GTP. Shown is a selection of cross-sectional images of Alexa 647-labeled FtsZ polymers inside GUIMVs. The image series begins on the outer wall of the GUV and progressively moves in the z-axis toward the opposite side of the vesicle through the GUV equator. The whole collection of images showed the same pattern (supplemental Fig. S3C). Scale bars = 10 μm.
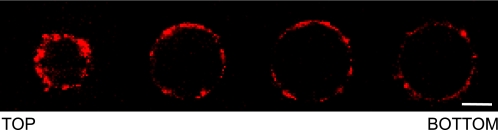
FtsA Localizes at the Inner Surface of GUIMVs
To visualize the incorporation and localization of FtsA inside GUIMVs, we obtained vesicles formed in the presence of FtsA labeled with Alexa 647 (Fig. 3). These confocal cross-sectional images revealed that FtsA was found in the vicinity of the inner surface of the vesicle. This observation was independent of the presence of ATP in the buffer. Moreover, in contrast with the observed distribution of GDP-FtsZ, the individual images collected throughout the whole volume of the GUV demonstrated that FtsA was not found dispersed in the vesicle lumen (supplemental Fig. S4).
FIGURE 3.
Spatial localization of Alexa 647-labeled FtsA inside GUIMVs in the absence of FtsZ. Shown are sequential cross-sectional images (taken every 6 μm) of Alexa 647-labeled FtsA inside GUIMVs. The whole collection of images (supplemental Fig. S4C) comprises the total volume of the spherical vesicle. Scale bar = 10 μm.
FtsZ Polymerization Has an Effect on FtsA Localization in GUIMVs
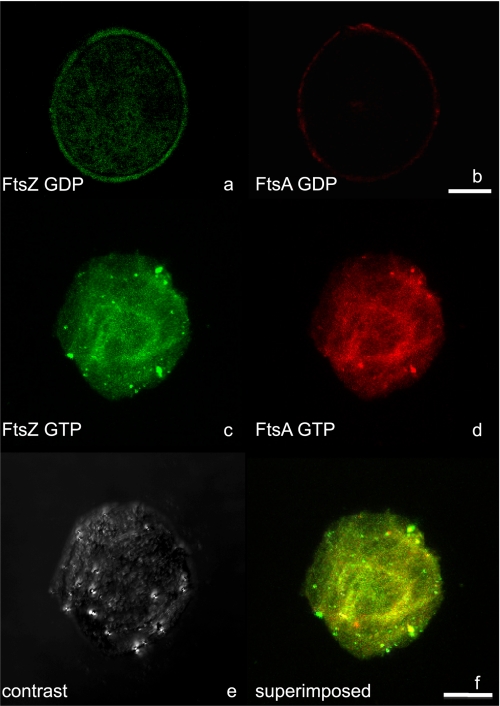
We decided to test the effect of FtsZ polymerization on the localization of FtsA within GUIMVs. Purified FtsA labeled with Alexa 647 (see “Experimental Procedures”) was found to decorate FtsZ polymers induced in the presence of GTP and Ficoll as crowding agent (supplemental Fig. S5) and therefore was capable of interacting in solution with FtsZ. In the presence of GDP-FtsZ, FtsA was found attached to the inner face of the vesicle membrane (Fig. 4b), as in the previous experiments without FtsZ. Most of the GDP-FtsZ was homogeneously distributed throughout the vesicle lumen; however, a faint signal of FtsZ was detected at the inner vesicle surface, indicating that a small fraction of FtsZ was bound to the membrane (Fig. 4a). It was expected that, under these conditions, FtsA, being attached to the membrane, would promote the polymerization of FtsZ at the inner surface of the vesicle. As shown in Fig. 4c, in the presence of GTP analogs (caged GTP induced by a UV laser), FtsZ was found polymerized as continuous fibers inside the GUIMVs. In this case, the structural organization changes of FtsZ polymers were accompanied by a redistribution of FtsA, which became dislodged from its peripheral localization at the inner side of the membrane and was preferentially localized, together with the FtsZ polymers, in the vesicle lumen (Fig. 4, d–f). The addition of 1 mm ATP did not alter the localization of FtsA inside the GUIMVs containing FtsZ.
FIGURE 4.
Spatial distribution/localization of Alexa 488-labeled FtsZ and Alexa 647-labeled FtsA inside the GUIMVs. a and b, image analysis of FtsZ and FtsA inside GUIMVs formed in the absence of GTP. a, equatorial cross-sectional image from a vesicle with Alexa 488-labeled FtsZ. b, equatorial cross-sectional image from the same vesicle with Alexa 647-labeled FtsA. c–f, image analysis of FtsZ and FtsA inside GUIMVs formed in the presence of 0.5 mm caged GTP. c and d, single equatorial sections of Alexa 488-labeled FtsZ and Alexa 647-labeled FtsA, respectively. e, differential interference contrast image. f, superposition of c and d. Scale bars = 10 μm.
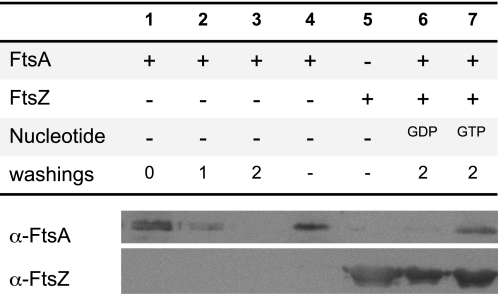
As the dissociation of FtsA from the inner membrane in the presence of FtsZ polymers was unexpected, an additional experiment to test the ability of FtsZ polymers to dislodge FtsA from the membrane was done. Fluorescently labeled FtsA and the inner membrane fractions used to prepare GUIMVs were mixed in solution without forming vesicles. Excess FtsA was then washed out by differential centrifugation. Sequential washings were performed until no traces of FtsA could be found in the wash supernatant (monitored by immunostaining) (Fig. 5, lane 3). No FtsA was detected in the supernatant when these membranes were treated with FtsZ and GDP (lane 6). However, a significant amount of FtsA was dislodged from the membrane fraction and appeared in the supernatant when GTP was added to the mixture (lane 7). From these experiments, we concluded that FtsA dissociates from the inner membrane to be localized with FtsZ polymers, in agreement with the previous observations made inside the GUIMVs.
FIGURE 5.
Immunoblot analysis of FtsA binding to inner membranes in the absence and in the presence of FtsZ. The membrane and soluble fractions were separated by differential centrifugation, and the presence of FtsZ and FtsA in these fractions was assayed with antibodies against both proteins as described under “Experimental Procedures.” Lanes 1–3, soluble fractions from samples of FtsA and inner membrane vesicles before a washing step (lane 1) and after one and two washing cycles (lanes 2 and 3, respectively; see “Experimental Procedures”); lanes 4 and 5, controls using purified FtsA and FtsZ proteins, respectively; lanes 6 and 7, soluble fractions from samples of FtsA and inner membrane vesicles previously incubated with either GDP-FtsZ (lane 6) or GTP-FtsZ (lane 7) polymers.
DISCUSSION
We have developed and used a novel synthetic approach to study the interaction and behavior of two proto-ring elements (FtsZ and FtsA) that play essential roles in bacterial division. For this purpose, micrometer-size natural membrane-bound vesicles (GUIMVs) were been obtained. These GUIMVs are structured systems in which the composition and environment are more similar to those found in the cell than in artificial lipid vesicles. These minimal acellular systems are amenable to biochemical and biophysical analyses of division events. Being contained systems, they allow mimicking the volume exclusion effects native to the bacterial interior by including crowding agents. These agents also favored the entrapment of proto-ring proteins inside the giant vesicles, a result that is in agreement with previous observations using other protein systems (33). Additionally, GUIMVs can be loaded with suitable nucleotides to trigger assembly reactions inside them.
FtsZ inside GUIMVs was a dispersed species in the presence of GDP. Irregular polymer networks located both at the lumen and in the vicinity of the inner vesicle surface were formed upon inclusion of GTP analogs. These results are consistent with earlier data obtained in the intact cell (34), indicating that most of the FtsZ molecules are located in the soluble cell fraction. Jones and Holland (35) observed that when the cell fractionation was done in the absence of exogenously added Mg2+, most of the FtsZ content was recovered in the cytoplasmic fraction, and only after extended sonic treatment in the presence of 10 mm Mg2+ was a significant fraction (as much as 40%) of FtsZ associated with the membrane fraction of maxicells. Because the physiological GTP/GDP ratio is high (36), FtsZ is likely to be found in the living cell as the GTP-bound form, mainly as FtsZ polymers. However, no Z-rings are observed during the early stages of the cell cycle (37), suggesting that, prior to Z-ring formation, the GTP-FtsZ species must remain in the cytoplasm. Moreover, from in vivo fluorescence recovery after photobleaching measurements using GFP-FtsZ, Stricker et al. (38) estimated that only 30% of the FtsZ molecules are located at the division ring.
Our finding that FtsA in isolation was localized in the vicinity of the inner surface of the GUIMVs is compatible with earlier work from Pla et al. (34) showing that FtsA was found in the cytoplasmic membrane upon cell fractionation. Yim et al. (19) observed that the removal of the C-terminal end of FtsA resulted in the loss of its biological function as judged by the inability to complement a nonsense mutation and by the failure to localize at the division ring. This C-terminal region of E. coli FtsA contains a membrane-targeting amphipathic helix (15), a domain that seems to be essential for FtsA function, as its deletion renders FtsA unable to localize at the membrane. We found that, in the absence of FtsZ, FtsA is associated with the inner surface of the GUIMVs, which suggests that FtsA is able to bind to the membrane before interacting with FtsZ (15).
A remarkable observation of our study was obtained when both FtsA and FtsZ were present inside the GUIMVs. In the presence of GDP, both proteins were localized at the same location as the individual isolated proteins (see above). However, the localization of FtsA was clearly different when FtsZ polymers were formed. Under these conditions, FtsA was dislodged from the membrane and found associated to the FtsZ polymers. This alternative localization of FtsA could have a regulatory role related to its amphitropic character; these proteins can be soluble or associated with the membrane depending upon certain conditions normally related to activation switches (39, 40). It has been previously shown that phosphorylation might regulate the association of FtsA with the inner membrane (14). It has also been reported that the membrane-associated region of FtsA can be replaced by the membrane motif of MinD, which is also an amphitropic protein (15). Further work will be needed to determine whether the displacement of FtsA from the inner membrane for incorporation into the FtsZ polymer might constitute a signal to activate some divisome function at late cell division stages or if it is a signal to indicate that the early stages of division have been completed.
Our finding was unexpected because it is generally thought that one of the roles of FtsA in cytokinesis is the attachment of FtsZ to the membrane (17). Accordingly to this proposed function for FtsA, we would expect that once FtsZ is allowed to polymerize inside the GUIMVs, the polymers would migrate toward the inner surface of the vesicle. We observed exactly the opposite behavior, with FtsZ polymers displacing the FtsA protein and themselves into the lumen of the vesicle. This behavior may indicate that the interaction between FtsZ and FtsA is stronger than the interaction between FtsA and the membrane.
In vivo, the interaction of FtsA and ZipA with FtsZ is required to assemble a functional Z-ring active in division. (If one of these proteins is absent, the ring formed by FtsZ is inactive (41).) In GUIMVs, even if ZipA is present, we found that FtsA does not retain FtsZ polymers in the vicinity of the membrane; on the contrary, FtsA becomes dislodged from its position when FtsZ polymerizes in the presence of GTP. This may be due to the amount of ZipA in the GUIMVs being insufficient to lock the other pair or, alternatively, to the interaction of FtsA with FtsZ being stronger than the interaction with the membrane. FtsA*, a variant form of FtsA (a hypermorph) in which the biological function of FtsA is mostly independent of the presence of ZipA (42, 43), has been described to modify the assembly of FtsZ polymers in vitro, causing their partial disassembly, therefore having a role similar to actin-depolymerizing proteins on F-actin (44). Our results suggest that in addition to its assumed role in mediating the association of FtsZ with the membrane at the time of division, FtsA may have other functions. Connections between FtsA and the late assembling divisome proteins seem to be the logical activities to be explored to search for those undescribed roles that may involve signaling of the completion of proto-ring assembly or, conversely, the activation of the late divisome functions. The use of GUIMVs will allow testing these alternatives, including the relative strength of the interactions between proto-ring components in a biochemically controlled acellular system, and will therefore contribute to the delineation of a more detailed picture of the function of the bacterial divisome.
Supplementary Material
Acknowledgments
We thank L. Rivas, J. R. Luque, F. Monroy, and I. López-Montero for advice and A. P. Minton, J. Mingorance, and B. Monterroso for useful comments.
This work was supported in part by European Commission DIVINOCELL FP7 HEALTH-F3-2009-223431, Ministerio de Ciencia e Innovación Plan Nacional BIO2008-04478-C03-00 (Spain), and Comunidad de Madrid COMBACT S-BIO-0260/2006 (to G. R. and M. V.).
M. J. dedicates this work to the memory of the late Amelia Partearroyo.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
A. Martos, B. Monterroso, B. Reija, S. Zorrilla, M. Vicente, G. Rivas, and M. Jiménez, manuscript in preparation.
B. Reija, B. Monterroso, M. Vicente, G. Rivas, and S. Zorrilla, manuscript in preparation.
- GUIMV
- giant unilamellar inner membrane vesicle
- GMPPCP
- guanosine 5′-(β,γ-methylene)triphosphate
- GUV
- giant unilamellar vesicle.
REFERENCES
- 1. Goehring N. W., Beckwith J. (2005) Curr. Biol. 15, R514–R526 [DOI] [PubMed] [Google Scholar]
- 2. Vicente M., Rico A. I. (2006) Mol. Microbiol. 61, 5–8 [DOI] [PubMed] [Google Scholar]
- 3. Vicente M., Rico A. I., Martínez-Arteaga R., Mingorance J. (2006) J. Bacteriol. 188, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Höltje J. V., Heidrich C. (2001) Biochimie 83, 103–108 [DOI] [PubMed] [Google Scholar]
- 5. Errington J., Daniel R. A., Scheffers D. J. (2003) Microbiol. Mol. Biol. Rev. 67, 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mingorance J., Rivas G., Vélez M., Gómez-Puertas P., Vicente M. (2010) Trends Microbiol. 18, 348–356 [DOI] [PubMed] [Google Scholar]
- 7. Nogales E., Downing K. H., Amos L. A., Löwe J. (1998) Nat. Struct. Biol. 5, 451–458 [DOI] [PubMed] [Google Scholar]
- 8. Mukherjee A., Dai K., Lutkenhaus J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Erickson H. P., Taylor D. W., Taylor K. A., Bramhill D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. González J. M., Vélez M., Jiménez M., Alfonso C., Schuck P., Mingorance J., Vicente M., Minton A. P., Rivas G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1895–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mingorance J., Tadros M., Vicente M., González J. M., Rivas G., Vélez M. (2005) J. Biol. Chem. 280, 20909–20914 [DOI] [PubMed] [Google Scholar]
- 12. Oliva M. A., Huecas S., Palacios J. M., Martín-Benito J., Valpuesta J. M., Andreu J. M. (2003) J. Biol. Chem. 278, 33562–33570 [DOI] [PubMed] [Google Scholar]
- 13. Bork P., Sander C., Valencia A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sánchez M., Valencia A., Ferrándiz M. J., Sander C., Vicente M. (1994) EMBO J. 13, 4919–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pichoff S., Lutkenhaus J. (2005) Mol. Microbiol. 55, 1722–1734 [DOI] [PubMed] [Google Scholar]
- 16. Hale C. A., de Boer P. A. (1997) Cell 88, 175–185 [DOI] [PubMed] [Google Scholar]
- 17. Pichoff S., Lutkenhaus J. (2002) EMBO J. 21, 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rivas G., López A., Mingorance J., Ferrándiz M. J., Zorrilla S., Minton A. P., Vicente M., Andreu J. M. (2000) J. Biol. Chem. 275, 11740–11749 [DOI] [PubMed] [Google Scholar]
- 19. Yim L., Vandenbussche G., Mingorance J., Rueda S., Casanova M., Ruysschaert J. M., Vicente M. (2000) J. Bacteriol. 182, 6366–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rueda S., Vicente M., Mingorance J. (2003) J. Bacteriol. 185, 3344–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. González J. M., Jiménez M., Vélez M., Mingorance J., Andreu J. M., Vicente M., Rivas G. (2003) J. Biol. Chem. 278, 37664–37671 [DOI] [PubMed] [Google Scholar]
- 22. De Vrije T., Tommassen J., De Kruijff B. (1987) Biochim. Biophys. Acta 900, 63–72 [DOI] [PubMed] [Google Scholar]
- 23. Osborn M. J., Gander J. E., Parisi E., Carson J. (1972) J. Biol. Chem. 247, 3962–3972 [PubMed] [Google Scholar]
- 24. Pott T., Bouvrais H., Méléard P. (2008) Chem. Phys. Lipids 154, 115–119 [DOI] [PubMed] [Google Scholar]
- 25. Montes L. R., Alonso A., Goñi F. M., Bagatolli L. A. (2007) Biophys. J. 93, 3548–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Méléard P., Bagatolli L. A., Pott T. (2009) Methods Enzymol. 465, 161–176 [DOI] [PubMed] [Google Scholar]
- 27. Rico A. I., García-Ovalle M., Mingorance J., Vicente M. (2004) Mol. Microbiol. 53, 1359–1371 [DOI] [PubMed] [Google Scholar]
- 28. Pla J., Sánchez M., Palacios P., Vicente M., Aldea M. (1991) Mol. Microbiol. 5, 1681–1686 [DOI] [PubMed] [Google Scholar]
- 29. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30. Zimmerman S. B., Trach S. O. (1991) J. Mol. Biol. 222, 599–620 [DOI] [PubMed] [Google Scholar]
- 31. Record M. T., Jr., Courtenay E. S., Cayley S., Guttman H. J. (1998) Trends Biochem. Sci. 23, 190–194 [DOI] [PubMed] [Google Scholar]
- 32. Altendorf K. H., Staehelin L. A. (1974) J. Bacteriol. 117, 888–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dominak L. M., Keating C. D. (2008) Langmuir 24, 13565–13571 [DOI] [PubMed] [Google Scholar]
- 34. Pla J., Dopazo A., Vicente M. (1990) J. Bacteriol. 172, 5097–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones C. A., Holland I. B. (1984) EMBO J. 3, 1181–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neurard J., Nygaard P. (1987) Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, pp. 445–473, American Society for Microbiology, Washington, D.C [Google Scholar]
- 37. Den Blaauwen T., Buddelmeijer N., Aarsman M. E., Hameete C. M., Nanninga N. (1999) J. Bacteriol. 181, 5167–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stricker J., Maddox P., Salmon E. D., Erickson H. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burn P. (1988) Trends Biochem. Sci. 13, 79–83 [DOI] [PubMed] [Google Scholar]
- 40. Johnson J. E., Cornell R. B. (1999) Mol. Membr. Biol. 16, 217–235 [DOI] [PubMed] [Google Scholar]
- 41. de Boer P. A. (2010) Curr. Opin. Microbiol. 13, 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geissler B., Elraheb D., Margolin W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geissler B., Shiomi D., Margolin W. (2007) Microbiology 153, 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beuria T. K., Mullapudi S., Mileykovskaya E., Sadasivam M., Dowhan W., Margolin W. (2009) J. Biol. Chem. 284, 14079–14086 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.