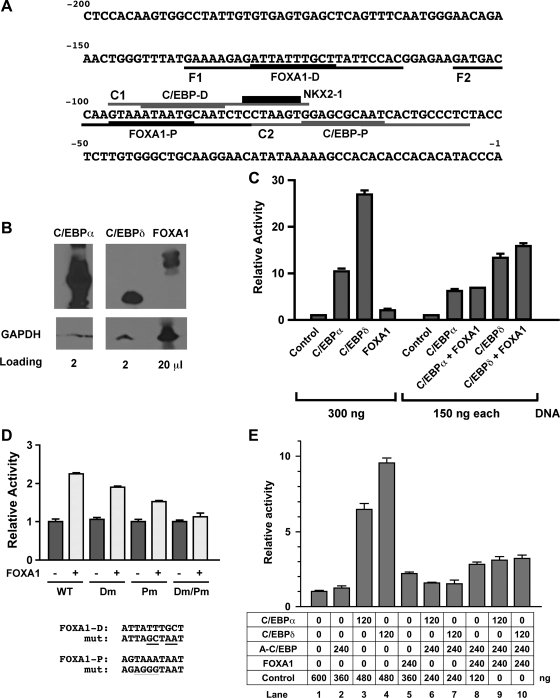
Fig. 4.
Luciferase reporter analysis of Scgb1a1 promoter activity. A: mouse Scgb1a1 gene promoter sequence up to −200 bp. Binding sites for transcription factors are as follows: C/EBP proximal (C/EBP-P), C/EBP distal (C/EBP-D), FOXA1 proximal (FOXA1-P), FOXA1 distal (FOXA1-D), and NKX2–1. Oligonucleotide sequences used for EMSA are F1, F2 containing FOXA1-D and FOXA1-P site, respectively, and C1, C2 containing C/EBP-D and C/EBP-P site, respectively. B: nuclear extracts were prepared from COS-1 cells transfected with C/EBPα, C/EBPδ, or FOXA1 expression plasmids and subjected to SDS-PAGE in duplicate, followed by Western blotting. After 1 membrane was cut into 4 sections containing only 1 loaded lane, each membrane was incubated with either anti-C/EBPα, C/EBPδ, or FOXA1. A duplicate blot was incubated with anti-GAPDH antibody that served as a loading control. The amount of samples loaded in each well is as follows: 2 μl for C/EBPα, 2 μl for C/EBPδ, and 20 μl for FOXA1. A gap was introduced between C/EBPα and C/EBPδ lanes because the original membrane contained another sample that was not included in this figure. C: COS-1 cells were cotransfected with the −457 Scgb1a1-luciferase reporter construct (300 ng) with various expression plasmids in the pcDNA 3.1 (vector) singly or in combination as indicated. D: COS-1 cells were cotransfected with −457 Scgb1a1-luciferase construct (300 ng) containing no (WT) or various FOXA1 binding site mutations as indicated together with (+) or without (−) FOXA1 expression plasmid (300 ng). Dm, FOXA1-D binding site mutant; Pm, FOXA1-P binding site mutant. Mutated (mut) sequences are shown at bottom. E: COS-1 cells were cotransfected with −457 Scgb1a1-luciferase reporter construct (300 ng) with various expression plasmids singly or in combination as indicated. Luciferase activity is mean ± SD of 3 independent experiments and is expressed based on that of the control vector as 1.