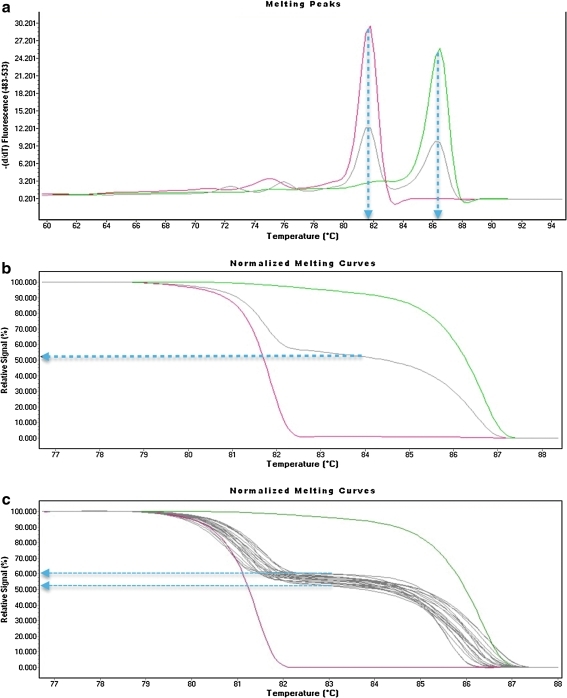
FIG. 1.
SNRPN quantitative HRM analysis in control samples. The colors indicate HRM analysis in AS deletion (red), PWS UPD (dark green), and a nonduplication control sample (gray). (a) Melting peaks indicate one peak at ∼81.7°C for the AS deletion sample that has no maternal allele and one peak at ∼86.2°C for PWS UPD that has no paternal allele. The normal control shows two peaks of approximate equal intensity at ∼81.7°C for the paternal allele and ∼86.2°C for the maternal allele. (b) The normalized melting profiles indicate that 100% of the relative signal intensity comes from the ∼81.7°C melting curve in the AS deletion sample, while 100% of the signal comes from the ∼86.2°C melting curve in the PWS UPD. A blue line drawn from the midpoint between the two melt curves in the nonduplication control sample indicates that ∼52% of the signal comes from the paternal melting curve (∼86.2°C) and 48% of the signal comes from the maternal melting curve (∼81.7°C). (c) A quantitative HRM analysis of multiple nonduplication control samples is shown, and although some variation in profiles could be detected, there was a very tight range of relative signal intensity in 15 nonduplication control samples between 52% and 60% (blue arrows). The individual melting curves ranged between 80.5°C and 82.3°C (average 81.4°C) for the paternal allele and 85°C–87°C (average 86°C) for the maternal allele. These values essentially match the peak intensity melt temperatures observed in (a) for the control. HRM, high-resolution melting; PWS/AS, Prader-Willi/Angelman syndrome; UPD, uniparental disomy.