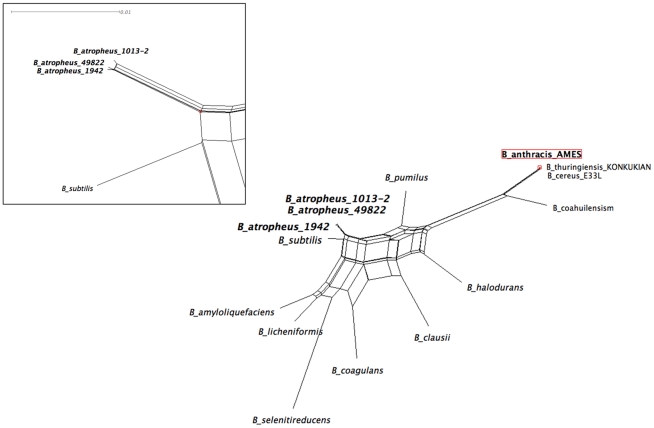
Figure 3. Identification of B. subtilis as nearest-neighbor to B. atrophaeus var. globigii by whole-genome phylogenetic analysis of Bacillus genomes.
Information-based genomic distance (IBGD) was determined by comparing the relative distributions of n-mers within each genome to generate a pair-wise matrix of relative n-mer frequencies (see Materials and Methods). Variation of the n-mer length between 4 and 8 did not substantially affect the derived phylogeny. In this case n-mer length of 5 was utilized. For clarity, only three select species of the B. cereus group (of more than 30 that all cluster together) are labeled on the figure. The apparent divergence of isolate 1013-2 is due to alteration of the n-mer frequencies as a result of the deletion of 72 kb of genomic material.