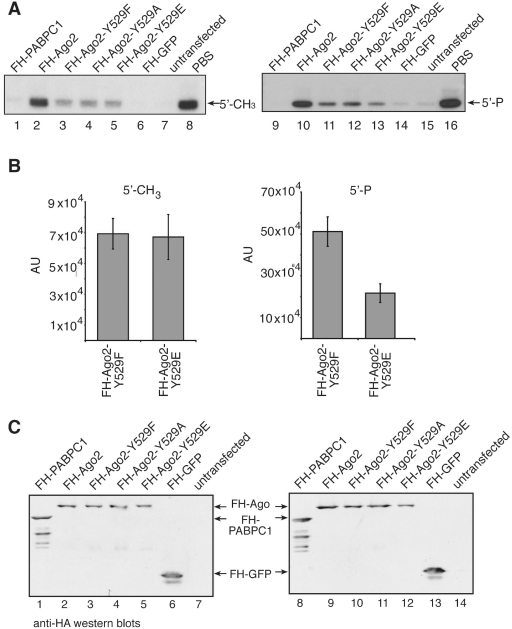
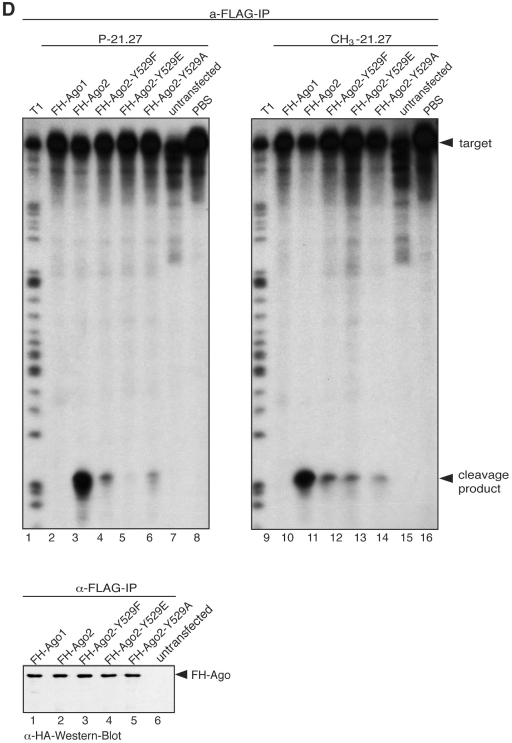
Figure 6.
Ago2 Y529 mutants bind 5′ methylated small RNAs equally well. (A) FH-PABPC1 (lanes 1 and 9) as an RNA-binding protein negative control, FH-Ago2 (lanes 2 and 10), FH-Ago2 Y529F (lanes 3 and 11), FH-Ago2 Y529A (lanes 4 and 12), FH-Ago2 Y529E (lanes 5 and 13) and FH-GFP as an unrelated negative control (lanes 6 and 14) were immunoprecipitated from HEK 293 cell lysates using anti-FLAG antibodies. Either a 5′ methylated (left panel) or 5′ phosphorylated (right panel) 21-nt single-stranded RNA was incorporated into the proteins. As control for unspecific degradation, PBS only was used (lanes 8 and 16). Tagged proteins were purified by immunoprecipitation and bound RNA was extracted for analysis by northern blotting using a probe specific to siR-21.27. (B) Quantification of the northern blot signals observed for Ago2 Y529F and Ago2 Y529E with a 5′ methylated (left panel) and a 5′ phosphorylated (right panel) 21-nt single-stranded RNA. Data used derive from two independent experiments. (C) Protein inputs that were used for the experiment shown in (A). (D) FH-Ago1 (lanes 2 and 10), FH-Ago2 (lanes 3 and 11), FH-Ago2 Y529F (lanes 4 and 12), FH-Ago2 Y529E (lanes 5 and 13), FH-Ago2 Y529A (lanes 6 and 14) were immunoprecipitated using anti-FLAG antibodies and subsequently incubated with a single stranded 5′ phosphorylated (lanes 2–8) or a 5′ methylated siRNA (lanes 10–16). After incubation a target RNA was added to each sample carrying a binding site of perfect complementarity to the siRNAs used. Cleavage products were analyzed by denaturing RNA PAGE. A western blot indicating the protein inputs is shown on the lower part of the figure.