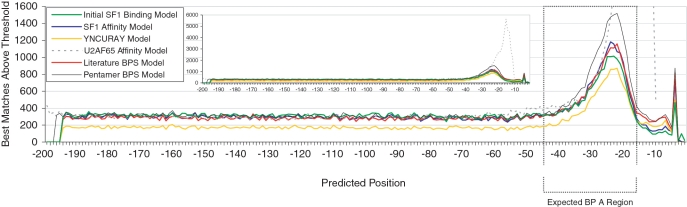
Figure 1.
Comparison of SF1 binding models to other profile models. Results of profile model searches in 117 499 human intronic regions (nucleotides from −199 to +1 relative to the 3′-splice site). For BPS models, the number of best matches above threshold (‘Materials and Methods’ section) was plotted based on the location of the predicted BP A. For the U2AF65 Affinity Model the number of best matches above threshold was plotted using the 5′-start position of the predicted PPT. The dotted box represents the expected BPS region (from −45 to −16). The area under the curve in the expected region, in excess of the number of background matches estimated from the number of matches at positions from −198 to −90, was used to evaluate and compare the models (‘Materials and Methods’ section). An unexpected sharp peak at position −4 was observed in all BPS models. This is the single position where the BP A does not conflict with the preferred pyrimidine preferences of the PPT. Indeed, a BP A has been mapped to position −4 of intron 3 of the human XPC gene, which also has a second BP A at position −24. Mutation of either BPS results in a variably penetrant form of familial xeroderma pigmentosum (18). The BPS with a BP A at position −24 is the top-scoring match for the SF1 Affinity Model while the BPS with BP A at position −4 also scores above threshold. The inset shows the distribution of profile matches with the y-axis scaled to display the complete profile match distribution for the U2AF65 Affinity Model.