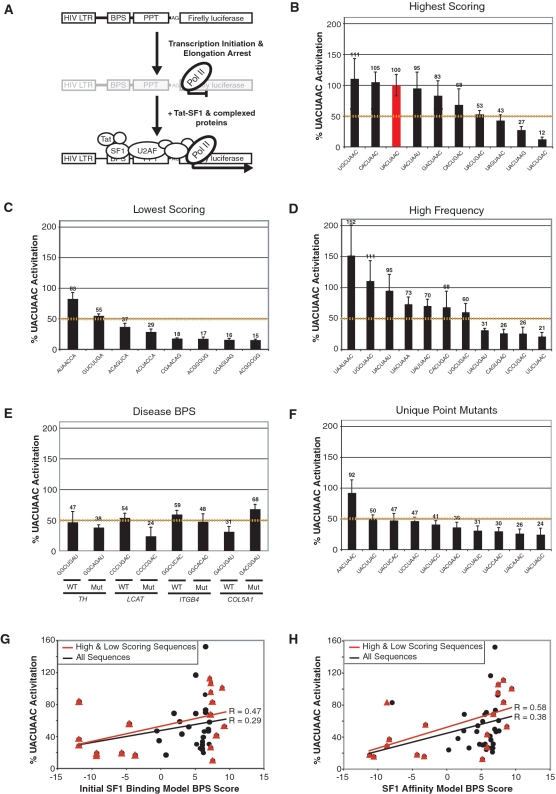
Figure 3.
SF1 binding to BPS reporters using the SF1 affinity Tat-hybrid assay. (A) Tat-hybrid system showing the HIV-1 FFL reporter engineered with the BPS, PPT and AG dinucleotide. In the absence of Tat-fused SF1, transcription elongation is arrested. Recruitment of the Tat fusion through the SF1-BPS and U2AF-RNA interactions enhances transcription elongation and luciferase expression. Panels (B–F) SF1 affinity data from Tat-hybrid assays for BPSs predicted by the Initial SF1 Binding Model; (B) 10 of the high-scoring sequences; (C) 8 very low-scoring sequences; (D) 11 sequences with high frequency rankings relative to all profile matches in the expected BPS region above a score threshold (‘Materials and Methods’ section); (E) wild-type and mutant BPS variants found in genetic diseases: TH—tyrosine hydroxylase intron 11; LCAT—lecithin cholesterol acetyltransferase intron 4; ITGB4—integrin beta-4 intron 31; COL5A1—collagen 5A1 intron 32; (F) the 10 remaining UACUAAC point mutants not included in A, B or C. The dashed yellow lines in panels B–F represent an arbitrary threshold for SF1 binding, set at 50% of activation relative to UACUAAC. (G) Correlation between bit score calculated using the Initial SF1 Binding Model and measured SF1 affinities (expressed as percent activation relative to UACUAAC) for the combined set of high and low scoring sequences as well as for all 46 sequences assayed. ‘R’ represents the correlation coefficient between bit score and SF1 affinity. (H) Correlation between bit score calculated using the SF1 Affinity Model and measured SF1 affinities as in (G).