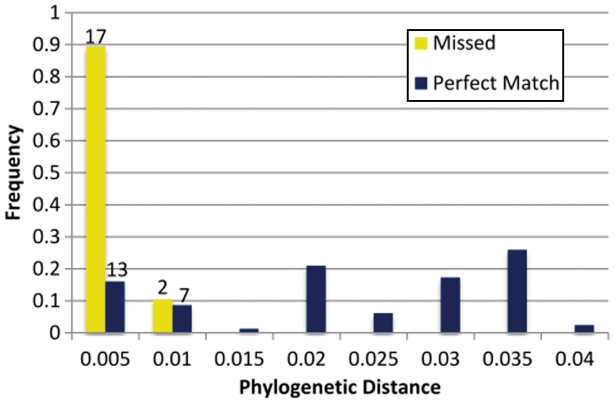
Figure 5.
Sensitivity of GiRaF as function of phylogenetic distance. Results from the ‘All Events’ dataset in Table 1, were categorized based on the F84 distance of implanted reassortments (from their original location) and the corresponding frequency histogram was graphed. This distance is a proxy for the sequence similarity of the (unobserved) ancestral sequences from which the two segments derived. GiRaF has nearly perfect sensitivity for implants with F84 distance >0.005 suggesting that the false positives are largely due to the challenge of distinguishing subtle events from phylogenetic noise.